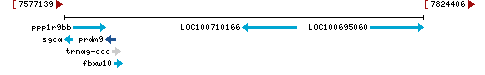- Location:
-
chromosome: LG8
- Exon count:
- 9
| Annotation release |
Status |
Assembly |
LG
|
Location |
|
104
|
current |
O_niloticus_UMD_NMBU (GCF_001858045.2) |
LG8 |
NC_031973.2 (7605708..7611968, complement) |
| 103 |
previous assembly |
ASM185804v2 (GCF_001858045.1) |
LG8 |
NC_031973.1 (2263479..2269764) |
Linkage Group LG8 - NC_031973.2