See "Essentials of Glycobiology, 4th Edition"
See the updated version of this chapter
Fungi are fascinating organisms that have been instrumental in defining the fundamental processes of glycosylation. This chapter describes the glycan structures of these diverse organisms and their synthesis, offers some insights into glycobiology revealed through studying fungal systems, and delineates the relationships of several important glycoconjugates to fungal biology and pathogenesis.
FUNGAL DIVERSITY
More than 70,000 species of fungi have been described, and it is likely that many more exist. The known species include yeast, molds, rusts, smuts, puffballs, and mushrooms. Study of these eukaryotes has been motivated by their unique and fascinating biology, their many useful products (including wine, cheese, and antibiotics), their utility as experimental systems for basic biology and protein expression, and their importance as animal and plant pathogens. Fungi are eukaryotic heterotrophs and absorb food from their environment. They are nonmotile and have life cycles that incorporate both sexual and asexual reproduction. They typically have elongated filaments or hyphae, which have cell walls that comprise complex polysaccharides including mannans, galactans, glucans, and chitin. There are four major phyla of fungi; each is extremely diverse, and fungi are assigned to a phylum on the basis of their mechanism for producing asexual spores. These phyla are the Chytridiomycota (primitive aquatic fungi), Zygomycota (e.g., black bread mold, Rhizopus nigricans), Ascomycota (sac fungi, e.g., Saccharomyces, Candida, Aspergillus, Neurospora, and morel mushrooms), and Basidiomycota (e.g., mushrooms, rot fungi, and puffballs). Some fungi are extremely beneficial to humans (e.g., the yeast Saccharomyces, which is used in fermentation) and some are not (e.g., pathogenic yeasts such as species of Candida and Crypotococcus).
YEAST AS A MODEL SYSTEM FOR BIOCHEMISTRY AND GLYCOBIOLOGY
More than 100 years ago, Louis Pasteur discovered that fermentation requires a viable organism, and since then yeast have been used as a model system to study cellular metabolism. In fact, Pasteur coined the word “ferment” during his work on alcohol production by yeast. Saccharomyces cerevisiae, or baker’s yeast, has been a wonderful resource for biologists and glycobiologists, especially because many of the fundamental enzymes in aerobic and anerobic metabolism (terms also invented by Pasteur) are shared between yeast and animals. Breakthroughs in enzymology occurred following the discovery by the Buchner brothers in 1897 that extracts of yeast could make ethanol and carbon dioxide from glucose, just like intact cells. Mannose is a major component of the yeast cell wall; it was discovered by Emil Fischer in 1888 and the mannose-rich glycans in yeast, historically called yeast gum, have been known since the 1890s. It is also noteworthy that the great chemist Sir Walter Norman Haworth discovered that the cell wall of yeast was composed of D-mannose. Haworth won the Nobel Prize in 1937 for his work on determining the chemical structures of carbohydrates (and vitamin C). The nucleotide sugar UDP-glucose was discovered by Luis Leloir in 1950 using yeast extracts, and he went on to discover GDP-mannose and other nucleotide sugars. Leloir won the Nobel Prize in chemistry in 1970 for this discovery of sugar nucleotides and their functions in carbohydrate synthesis. Studies with yeast were also important in defining the dolichol-linked sugars and their roles as intermediates in protein N-glycosylation and in elucidating numerous other fundamental biochemical pathways including the synthesis of sterols from acetate and fatty acid oxidation.
Yeast cells have a nucleus, a nuclear envelope associated with the endoplasmic reticulum (ER), a Golgi apparatus, mitochondria, and peroxisomes. These simple eukaryotes provide both a convenient source of enzymes for study and a powerful genetic system. Yeast secretory mutants (Sec mutants) helped to define the components of the protein secretory pathway, by which proteins travel from the ER to the cell surface and the exterior of the cell, becoming glycosylated en route. Yeast cells with various mutations have also been instrumental in elucidating mechanisms and pathways of protein N-glycosylation and steps in the biosynthesis of glycosylphosphatidylinositol (GPI)-anchored glycoproteins. At the same time, studies in yeast have certain limitations. These cells do not synthesize typical complex N-glycans, mucins or mucin-type O-glycans, O-linked N-acetylglucosamine (O-GlcNAc), or glycosaminoglycans of the types found in vertebrates. In addition, although yeast are valuable for studying sphingosine and sphingolipid metabolism, they do not synthesize complex glycosphingolipids or gangliosides like those found in mammals. However, they do make some very unusual glycolipids, including short, mannose-containing compounds. Finally, most fungi do not synthesize sialic acid, have relatively low amounts of fucose, and lack long-chain glycolipids (apart from those glycolipids participating in the synthesis of GPI anchors or glycoproteins in the ER).
GLYCAN STRUCTURES OF YEAST
All fungi have cell walls, which are critical to maintaining cell shape and integrity in environments that range from the surface of grapes to human tissues. Cell walls are highly cross-linked structures, which adapt to growth conditions in a dynamic and flexible way (Fig. 21.1). The cell-wall polysaccharides that have been described so far are composed of polymers of mannose, glucose, galactose, N-acetylglucosamine, and/or rhamnose, and these include mannans, glucans, chitin, galactomannans, glucomannans, rhamnomannans, and phosphomannans. In mannans, the mannose residues of the polymeric backbone are α-linked (usually α1–6), whereas in glucans, the glucose residues are β-linked (mostly β1–3, although some are β1–6). The interconnected polymers may have other sugars attached to these backbone structures and additional modifications that are specific to each organism. Fungal cell walls also contain covalently and noncovalently linked glycoproteins that bear N- and O-glycans of myriad structures; some of these glycoproteins begin as GPI-anchored proteins.
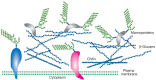
FIGURE 21.1
Illustration of the cell wall of fungi, showing the presence of glycoproteins and mannoproteins in the layer of the wall and an inner layer of different polysaccharides. The presence of different types of glucans and chitin varies between different fungal (more...)
Chitin is a polymer of β1–4-linked GlcNAc, which occurs in chains that typically exceed 1000 residues. These chains self-associate to form microfibrils and are deposited primarily at the bud neck of yeast or at septa in filamentous fungi. Chitin synthesis is highly regulated by the coordinated action of multiple chitin synthases, so that deposition occurs at the specific sites and times required for normal cell growth and division. In S. cerevisiae three chitin synthases (Chs1p, Chs2p, and Chs3p) have been described, along with multiple additional gene products that participate in the regulation and localization of chitin synthesis. Chitin may also be deacetylated to form another polymer, chitosan. In S. cerevisiae this process occurs during spore formation, although in other fungi chitosan is required for cell wall functions during vegetative growth. Chitin is also the primary component of exoskeletons of arthropods (see Chapter 24) and in the cuticle of nematodes (see Chapter 23).
Yeast express a relatively simple array of glycolipids, although Candida albicans is notable for its large lipid-linked mannans. Many fungi make short-chain glycolipids, commonly containing myoinositol phosphate linkers to mannose that may be modified by galactofuranose (as in Histoplasma capsulatum) or an additional mannose residue. Some longer galactose- and mannose-containing glycolipids are found in Aspergillus niger. Short-chain glycosylceramides such as Glc-Cer and Gal-Cer are also found in the fungi Schizophyllum commune and Aspergillus fumigatus, respectively. Examples of fungal species are highlighted below to illustrate the many general glycan features that yeast share while emphasizing the glycoconjugates that are unique to each organism. The last two sections of this chapter provide notable fungal examples of glycoconjugate biosynthesis.
Saccharomyces cerevisiae, the Model Yeast
S. cerevisiae, by far the best-studied fungal organism, is a round budding yeast approximately 5–10 μm in diameter. This single-celled eukaryote contains an amazing assortment of different glycoconjugates, including proteins with N- and O-glycan modifications, polypeptides bearing mannan polysaccharides (Figures 21.2 and 21.3a), GPI-anchored glycoproteins (Figure 21.4), glycolipids, and cell-wall polymers. The O-glycans have core mannose residues linked to Ser/Thr and the N-glycans are all linked to Asn residues in the sequon -Asn-X-Ser/Thr-, where X can be any amino acid except proline. S. cerevisiae has few glycolipid components in the outer membrane and the best characterized are small phytoceramide derivatives of myo-inositol phosphate that contain a single residue of mannose (mannose inositol-phosphate-ceramide [MIPC] and M(IP)2C). Interestingly, no galactose-containing glycans have been described in S. cerevisiae, although glycans with galactose are expressed by other fungi.
FIGURE 21.2
Structures of selected yeast mannans. Note that a single pyruvate is (R)4,6 acetyl-(ketal)-linked to the terminal galactose residue in the pyruvylated structure. Symbol Key:
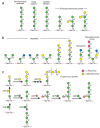
FIGURE 21.3
Structures of selected O-linked glycans in fungi: (a) yeast, (b) Aspergillus, and (c) Cryptococuus. Symbol Key:

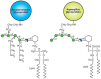
FIGURE 21.4
Structures of two yeast glycosylphosphatidylinositol (GPI)-anchors. Hexagon indicates myo-D-inositol. Symbol Key:

GPI-anchored glycoproteins of S. cerevisiae have a conserved trimannose core linked to glucosamine-inositol-phosphatide, substituted with an additional α1–2-linked mannose and ethanolamine phosphodiesters (Figure 21.4) (see Chapter 11). The biosynthesis of these structures is complex, involving more than 20 genes, and it proceeds by the formation of a lipid precursor whose synthesis involves many of the same steps as those found in the synthesis of mammalian GPI anchors. The GPI precursor is incorporated near the carboxyl terminus of nascent glycoproteins in the ER by a GPI transamidase, which is directed to the site of addition by a signal sequence that is concomitantly cleaved. In an interesting divergence from higher eukaryotes, the GPI anchor is partly removed from some yeast glycoproteins, and the remaining glycosyl portion of the GPI structure, with the attached protein, becomes covalently linked to cell-wall β1–6 glucans.
The cell wall of S. cerevisiae is a formidable structure and may account for up to one third of the dry weight of the cell. The major components of this cell wall are membrane-associated glycoproteins (mannoproteins) and β-linked glucans, along with some chitin that is primarily in the bud scars. The wall is a layered structure, with the rigid β-glucans proximal to the membrane surrounded by linked mannoproteins and additional β-glucans to form an elastic network. Cell-wall mannoproteins have the conserved N-glycan core structure linked to Asn residues, but this structure is further elaborated with an extensive repeating α1–6-linked mannose chain. The repeating α1–6-linked mannose backbone is usually branched by short chains of α1–2- and α1–3-linked mannose structures; some of these side chains may be in phosphodiester linkage (Figure 21.2). The mannans are highly heterogeneous in length and branching. In addition to large N-glycans rich in mannose, yeast cell walls contain proteins bearing Ser/Thr-linked O-mannose glycans, although these structures are of modest size compared to the mannans on N-glycans (Figure 21.3a). As mentioned above, many of the cell-wall mannoproteins in yeast also originate on GPI anchors (Figure 21.4). A wide assortment of mutants in mannan biosynthesis (termed mnn mutants) have been identified in S. cerevisiae, and a significant percentage of the genome (up to 20%) may be involved in elaborating the cell wall and its varied components.
Schizosaccharomyces pombe, a Fission Yeast
S. pombe is a rod-shaped fission yeast, which is 3–4 μm in diameter and 7–14 μm in length. Rather than budding, this organism grows by elongation and fission to give equal-sized daughter cells. Like S. cerevisiae, it has a relatively small genome of approximately 14 million base pairs. S. pombe has been useful as a genetically manipulatable model organism for studying the cell cycle. Because it has well-defined organelle structures compared to other yeast, it is a popular choice for studies of intracellular structure. S. pombe also synthesizes mannoproteins and mannans, some containing caps of α1–2-linked galactose residues that may also be pyruvylated. The galactose residues are important in lectin recognition in non-sexual flocculation (clumping) of S. pombe, as evidenced by inhibition of this process by free galactose. In contrast, flocculation in S. cerevisiae is mannose-dependent and inhibited by free mannose. The newly synthesized N-glycans in both S. cerevisiae and S. pombe have nine mannose residues (Man9GlcNAc2Asn; Figure 21.5). In S. cerevisiae, these structures are trimmed in the ER to form Man8GlcNAc2Asn (as discussed below), but this does not occur in S. pombe. In contrast to the model yeast, some S. pombe O-linked mannose structures contain galactose (Figure 21.3a).
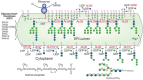
FIGURE 21.5
Biosynthesis of N-glycans and addition of N-glycans to -Asn-X-Ser/Thr- residues in newly synthesized glycoproteins in the yeast endoplasmic reticulum (ER). Individual steps in the biosynthesis pathway from dolichol phosphate and the structure of dolichol (more...)
Pichia pastoris, a Popular Expression System
P. pastoris is a methylotrophic, nonpathogenic organism that was discovered in 1969 in a screen for yeast capable of utilizing methanol. Methanol is oxidized to formaldehyde and hydrogen peroxide by alcohol oxidase (AOX) in the peroxisome. The formaldehyde exits the peroxisome and is oxidized to formate and carbon dioxide in the cytoplasm for the purpose of energy production. Any remaining formaldehyde is assimilated into glyceralde-hyde-3-phosphate and dihydroxyacetone by condensation with xylulose-5-monophos-phate in a reaction catalyzed by the peroxisomal enzyme diydroxyacetone synthase. P. pastoris has become popular as a model system for making recombinant proteins because it is easy to manipulate genetically and can be grown to very high densities. The promoter for AOX is methanol inducible, and transcripts driven by this promoter may comprise up to 5% of the total poly(A)+ RNA in induced cells.
P. pastoris has several advantages over Escherichia coli as an expression system in that it does not produce inclusion bodies and it promotes the correct folding of eukaryotic proteins. It also has certain advantages over yeast. First, although its basic pathway of N-glycosylation is similar to that of S. cerevisiae and yields glycoproteins with oligomannose-type N-glycans, these structures in P. pastoris have only 5–15 mannose residues (typically Man9GlcNAc2Asn and Man8GlcNAc2Asn) compared with the 50–150 mannose residues found in glycoproteins from S. cerevisiae (see Figure 21.2). Hyperglycosylation in S. cerevisiae can interfere with protein folding, requiring use of mnn mutants to limit hyperglycosylation and avoid this problem. Also, P. pastoris does not add outer α1–3-linked mannose residues to its N-glycans. These structures are highly antigenic to humans, making proteins expressed in S. cerevisiae unsuitable for human pharmaceutical use. P. pastoris synthesizes O-glycans with an O-linked mannose core attached to Ser/Thr residues; most of these are short α1–2-linked mannose structures (see Figure 21.3a). Recently, P. pastoris has been genetically modified to produce therapeutic glycoproteins with humanized glycosylation.
Kluyveromyces lactis, a Yeast of Industrial Interest
K. lactis metabolizes lactose to lactic acid and, along with A. niger and E. coli, it is grown to produce rennet for making cheese and other products. K. lactis is also a rich source of β-galactosidase, which hydrolyzes lactose. K. lactis synthesizes mannans similar to those in S. cerevisiae, but they lack mannose phosphate modifications, and some side chains are capped with a residue of N-acetylglucosamine. A K. lactis mutant that lacks these N-acetyl-glucosamine residues because of a deficiency of the Golgi UDP-GlcNAc nucleotide sugar transporter has been productively exploited in studies of heterologous transporters.
Cryptococcus albidus and C. laurentii, Two Environmental Fungi
Cryptococcus is an encapsulated basidiomycete found in all environments. There are nearly 40 known species of Cryptococcus, including the pathogen C. neoformans (which is discussed below). Two common species of Cryptococcus that are not pathogenic are C. albidus and C. laurentii. The C. albidus capsule is composed of polysaccharides with a backbone of α1–3-linked mannose residues, some of which are O-acetylated and/or substituted with β1–2-linked xylose or glucuronic acid residues, similar to the glucuronoxylomannans in C. neoformans. C. laurentii is an encapsulated yeast that is prevalent in the tundra and it has the unusual property of producing toxins that kill the yeast Candida albicans. The O-glycans of C. laurentii have been well studied and are unusual in that they contain mannose, xylose, and galactose (Figure 21.3c) and they are synthesized by a unique set of mannosyl-, xylosyl-, and galactosyltransferases that are not homologous to human enzymes.
Pseudallescheria boydii, an Emerging Pathogen
P. boydii is found in soils and polluted water and is an emerging pathogen in immunocompromised individuals. The mycelial form of P. boydii synthesizes a novel peptidorhamnomannan that contains both N- and O-glycans. It is characterized by immunodominant α 1–3-linked rhamnopyranose caps on the glycans, as shown in Figure 21.3b.
Candida albicans, an Important Pathogenic Yeast
C. albicans is a normal commensal organism that under various circumstances causes illness ranging from irritations of mucosal surfaces to life-threatening systemic infections. The Candida cell wall consists of mannans similar to those in S. cerevisiae, but they are termed phosphopeptidomannans. It also produces short β1–2-linked mannose glycans that are highly antigenic (Figure 21.2). These unusual β1–2-linked mannose glycans are also expressed on phospholipomannan antigens (PLMs). PLMs contain phytoceramide derivatives of myo-inositol phosphate to which are linked mannose and long polysaccharides of β1–2-linked mannose. The cell wall also contains β1–3- and β1–6-linked glucans and chitin. The O-glycans of C. albicans are short mannose-containing chains that have α1–2-linked mannose (Figure 21.3a) but lack the α1–3-linked mannose caps found in S. cerevisiae. As in S. cerevisiae, deficiencies of O-mannose addition generated through genetic deletions are lethal, indicating that O-mannosylation is essential in this yeast. C. albicans mannans are also important in its interactions with host cells, including macrophages and dendritic cells. In particular, these structures are recognized by the mannose receptor and by dectin-2. These are C-type lectins expressed by immune cells and they are important in both innate and adaptive immune responses (see Chapter 31). PLMs may be shed by C. albicans, and through interactions with Toll receptors (TLR-2), they can induce NF-κB activation and cytokine responses such as tumor necrosis factor-α (TNF-α) secretion. Galectin-3, a ubiquitous member of the galectin family of lectins that is highly expressed in macrophages, also appears to recognize C. albicans expressing β1–2-linked mannose residues, resulting in opsonization of the yeast.
Dictyonema glabratum Adopts a Symbiotic Lifestyle
Basidiomycetes are fungi that produce spores formed from a pedestal-like structure called the basidium. Examples of basidiomycetes include fungi that have gills or pores, such as common mushrooms and bracket fungi as well as the Cryptococcus species discussed above. D. glabratum illustrates another fungal lifestyle as this organism lives in symbiosis with cyanobacteria Scytonema sp. forming a lichen. This lichen synthesizes many glycans that are unusual compared to those of other lichen and fungi, including β-glucans, mannans, and xylans. For example, the β-glucans of most lichens are linear, but in D. glabratum they are branched with β1–3 and β1–6 linkages. The mannans of D. glabratum have an α1–3-linked backbone, rather than the typical α1–6 linkages found in other lichens, along with branches at the 2 and 4 positions. Finally, the xylans of this organism are linear β1–4-linked polymers of xylose, more typical of those found in higher plants and algae than in fungi. D. glabratum also synthesizes some unusual short glycolipids, including glycosyldiacylglycerolipids, which are similar to plant glycolipids and contain mono-, di-, and linear trisaccharides of α1,6-linked galactopyranose. This fungus thus serves as one more example of the extensive and surprising diversity seen in the fungal kingdom.
PROTEIN GLYCOSYLATION IN S. CEREVISIAE
The yeast S. cerevisiae has been an excellent model system for the study of certain glycan synthetic pathways. Many successful studies have used various approaches to select for glycosylation-deficient mutants. For example, mannan biosynthesis was partly defined by selecting mutants for altered antibody binding. Other important mutants that have proven valuable in studying N-glycosylation pathways were generated by [3H]mannose suicide selection; only cells that incorporate unusually low levels of [3H]mannose survive growth in the presence of this compound. The survivors included yeast mutants that were defective in N-glycosylation, termed the alg mutants for their deficiency in asparagine-linked glycosylation. The alg mutants were sorted into complementation groups and found to be defective in adding glycans to polypeptide precursors, thus explaining the reduced mannose incorporation. At nonpermissive temperatures, these mutants accumulated specific lipid-linked oligosaccharides immediately upstream of the defective biosynthetic reactions. Analysis of the accumulated intermediates allowed correlation of the mutants with steps in the assembly of the dolichol-P-P-oligosaccharide precursor (Figure 21.5) and identification of the enzymes involved. Mammalian cells have homologs to each of these yeast enzymes and they act to synthesize the conserved lipid-linked core glycan donor, Glc3Man9GlcNAc2-P-P-Dol, which is transferred to nascent polypeptides in the ER (see Chapter 8).
Following core N-glycosylation, the Glc3Man9GlcNAc2Asn-R is processed in mammals and yeast, with removal of glucose residues by α-glucosidases I and II to generate Man9GlcNAc2Asn-R. In mammals andS. cerevisiae, the Man9GlcNAc2Asn-R is further trimmed to Man8GlcNAc2 Asn-R by an ER-mannosidase. S. pombe lacks this enzyme and stops processing at Man9GlcNAc2Asn-R. The Man8GlcNAc2Asn-R in mammals and the Man9GlcNAc2Asn-R in S. pombe are substrates for the UDP-Glc:glycoprotein glucosyltransferase that generates Glc1Man8GlcNAc2Asn and Glc1Man9GlcNAc2Asn in mammals and S.pombe, respectively. This reglucosylation is part of the quality control system in protein folding in the ER (see Chapter 36). The monoglucosylated structure is a ligand for the chaperone lectins calnexin and calreticulin in mammalian cells. Although S. cerevisiae lacks this glucosyl-transferase, both S. cerevisiae and S. pombe express calnexin, but lack a calreticulin homolog. The newly synthesized S. cerevisiae glycoproteins may acquire monoglucosylated, oligomannose N-glycans during partial deglucosylation by α-glucosidase II, which may be sufficient to allow them to interact with the quality-control pathway. However, the quality-control system in S. cerevisiae may also be weak compared to that of S. pombe and mammalian cells, because the survival of S. cerevisiae is not affected by mutations causing loss of the calnexin homolog.
Yeast protein O-linked mannose is added by ER protein mannosyltransferases (PMTs), which use Dol-P-Man as a mannose donor. There are several PMTs in S. cerevisiae and each may have a different substrate specificity and glycoprotein preference. Mammals also have protein O-mannosyltransferases (POMTs) that use Man-P-Dol as a donor to generate Manα-Ser/Thr-modified glycoproteins in the ER (see Chapter 12). The Man-P-Dol for the yeast PMTs is synthesized as shown in Figure 21.5 (top right) and is used for both N- and O-glycosylation pathways. Subsequent additions of mannose residues to growing chains occur in the Golgi apparatus, where GDP-Man serves as the donor for reactions catalyzed by Mn++-dependent mannosyltransferases. This is interesting in topological terms, because GDP-Man is used to generate both the biosynthetic Man5GlcNAc2-P-P-Dol precursor, which is then “flipped” into the ER, and the Dol-P-Man donor of additional mannosylation reactions (Figure 21.5). Thus, GDP-Man does not appear to be transported into the ER, although it is transported into the Golgi apparatus.
Mannan extensions are generated in the Golgi apparatus by mannosyltransferases that utilize GDP-Man as the donor, with one or more specific glycosyltransferases to catalyze synthesis of each linkage and branch. Phosphomannose addition occurs by transfer of Man-1-P from GDP-Man donors in the Golgi apparatus. Many mutants in mannan biosynthesis (mnn) have been identified that lead to truncated mannans.
CAPSULE BIOSYNTHESIS IN THE PATHOGEN CRYPTOCOCCUS NEOFORMANS
C. neoformans is a common soil-dwelling, encapsulated fungus. It latently infects healthy people but causes severe disease in immunocompromised individuals and is an AIDS-defining illness. C. neoformans is unique among pathogenic fungi in having an extensive polysaccharide capsule that is required for its virulence (Figure 21.6). Two major capsular polysaccharides named for their monosaccharide components are glucuronoxylomannan (GXM) and a galactoxylomannan (GalXM). GXM is an extended α1–3 mannan substituted with β1–2Xyl, β1–4Xyl, and β1–2GlcA, and a subset of the mannose residues are 6-O-acetylated. Several serotypes of C. neoformans differ in the xylose modifications of the repeating unit; serotype B is shown in Figure 21.7. The second polymer, GalXM, is based on α1–6 galactan, with side chains of galactose, mannose, and xylose (Figure 21.7). In GalXM, most of the galactose is galactopyranose, although galactofuranose has been reported in this structure. The cryptococcal galactofuranose donor is generated by the enzyme UDP-galactopyranose mutase. Molecularly, GXM is substantially larger than GalXM (1.7–7.4 ×106 daltons for GXM vs. 105 daltons for GalXM).

FIGURE 21.6
A quick-freeze deep-etch image of the edge of a C. neoformans cell. The polysaccharide capsule (open meshwork at right) is linked to the cell wall (central structure dividing the image from upper left to lower right) via α1–3 glucan. The (more...)
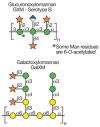
FIGURE 21.7
Structures of capsular polysaccharides in Cryptococcus neoformans. Symbol Key:

The capsule is a dynamic structure that can change in thickness and composition depending on the environment and growth conditions. Under standard in vitro conditions, the capsule is approximately 1–2 μm in diameter, but it can be much thicker, especially in the context of mammalian infection. Association of the capsule with the cell surface relies on a cell-wall component, α1–3 glucan. Although α1–3 glucan is not present in the cell walls of S. cerevisiae or C. albicans, it is common in other fungi.
Cell biological studies indicate that cryptococcal capsule components are generated intracellularly. Investigations of capsule biosynthesis have taken both molecular and biochemical approaches. For example, mutants in capsule structure have been identified through genetic screening approaches based on colony morphology and antibody reactivity, allowing identification of genes required for capsule synthesis. Genome analysis has also identified candidate biosynthetic enzymes. Finally, direct biochemical studies of cryptococcal enzymes have been applied to this question. Together, these approaches have begun to elucidate the biosynthetic pathways required to generate this elaborate and important structure.
FURTHER READING
- Spencer JF, Gorin PA. Mannose-containing polysaccharides of yeasts. Biotechnol Bioeng. 1973;15:1–12. [PubMed: 4123390]
- Ballou CE. Some aspects of the structure, immunochemistry, and genetic control of yeast mannans. Adv Enzymol Relat Areas Mol Biol. 1974;40:239–270. [PubMed: 4599414]
- Ballou CE, Raschke WC. Polymorphism of the somatic antigen of yeast. Science. 1974;184:127–134. [PubMed: 4131378]
- Ballou CE, Lipke PN, Raschke WC. Structure and immunochemistry of the cell wall mannans from Saccharomyces chevalieri, Saccharomyces italicus, Saccharomyces diastaticus, and Saccharomyces carlsbergensis. J Bacteriol. 1974;117:461–467. [PMC free article: PMC285535] [PubMed: 4590470]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci. 1983;80:7466–7470. [PMC free article: PMC389972] [PubMed: 6369318]
- Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993;7:540–550. [PubMed: 8472892]
- Bretthauer RK, Castellino FJ. Glycosylation of Pichia pastoris-derived proteins. Biotechnol Appl Biochem. 1999;30:193–200. [PubMed: 10574687]
- Burda P, Jakob CA, Beinhauer J, Hegemann JH, Aebi M. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology. 1999;9:617–625. [PubMed: 10336995]
- Dickson RC, Lester RL. Yeast sphingolipids. Biochim. Biophys. Acta. 1999;1426:347–357. [PubMed: 9878820]
- Gemmill TR, Trimble RB. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta. 1999;1426:227–237. [PubMed: 9878752]
- Herscovics A. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1999;1426:275–285. [PubMed: 9878780]
- Parodi AJ. Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells. Biochim. Biophys. Acta. 1999;1426:287–295. [PubMed: 9878790]
- Calderone R, Suzuki S, Cannon R, Cho T, Boyd D, Calera J, Chibana H, Herman D, Holmes A, Jeng HW, Kaminishi H, Matsumoto T, Mikami T, O’Sullivan JM, Sudoh M, Suzuki M, Nakashima Y, Tanaka T, Tompkins GR, Watanabe T. Candida albicans: Adherence, signaling and virulence. Med. Mycol. 2000;38(suppl 1):125–137. [PubMed: 11204138]
- Bencurova M, Rendic D, Fabini G, Kopecky EM, Altmann F, Wilson IB. Expression of eukaryotic glycosyltransferases in the yeast Pichia pastoris. Biochimie. 2003;85:413–422. [PubMed: 12770780]
- Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. A yeast under cover: The capsule of Cryptococcus neoformans. Eukaryot Cell. 2003;2:655–663. [PMC free article: PMC178345] [PubMed: 12912884]
- Hamilton JF, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, Wildt S, Gerngross TU. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. [PubMed: 12947202]
- Poulain D, Jouault T. Candida albicans cell wall glycans, host receptors and responses: Elements for a decisive crosstalk. Curr Opin Microbiol. 2004;7:342–349. [PubMed: 15358252]
- Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: A useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–138. [PubMed: 15565717]
- Wildt S, Gerngross TU. The humanization of N-glycosylation pathways in yeast. Nat Rev Microbiol. 2005;3:119–128. [PubMed: 15685223]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. [PubMed: 16317064]
- Klutts JS, Yoneda A, Reilly MC, Bose I, Doering TL. Glycosyltransferases and their products: Cryptococcal variations on fungal themes. FEMS Yeast Res. 2006;6:499–512. [PubMed: 16696646]
- Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia. 2006;98:838–849. [PubMed: 17486961]
- Klis FM, Ram AF, De Groot PW. A molecular and genomic view of the fungal cell wall. The Mycota VIII. In: Howard RJ, Gow NAR, editors. Biology of the fungal cell. 2. Springer-Verlag; Berlin: 2007. pp. 97–120.
- Schutzbach J, Ankel H, Brockhausen I. Synthesis of cell envelope glycoproteins of Cryptococcus laurentii. Carbohydr Res. 2007;342:881–893. [PMC free article: PMC2600673] [PubMed: 17316583]
Publication Details
Author Information and Affiliations
Authors
Richard D Cummings and Tamara L Doering.Copyright
Publisher
Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY)
NLM Citation
Cummings RD, Doering TL. Fungi. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 21.






