NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.
The primary goal of the project was the identification of chemical probes decreasing the storage lipid content of cells. For this purpose, a cell-based assay using embryonic Drosophila melanogaster cells called “S3” was developed. Cells were incubated with the free fatty acid oleate to induce storage lipid deposition. Lipids are stored in specialized organelles, the so-called lipid droplets, which were stained with the lipophilic dye BODIPY493/503 (Invitrogen/Molecular Probes). We identified three unique chemotypes showing potent lipid droplet reduction phenotypes in the S3 cell based assay. The three probes identified, ML206, ML219 and ML220, had EC50s of 8, 2 and 705 nM, respectively. While ML206 showed little translation into mammalian cell lines, both ML219 and ML220 showed moderate translation into acute myeloid leukemia (AML)12 cells.
Assigned Assay Grant #: R03MH085686
Screening Center Name & PI: NIH Chemical Genomics Center, Christopher P. Austin
Chemistry Center Name & PI: NIH Chemical Genomics Center, Christopher P. Austin
Assay Submitter & Institution: Dr. Mathias Beller, Max-Planck-Institut für Biophysikalische Chemie
PubChem Summary Bioassay Identifier (AID): 1623
Probe Structure & Characteristics
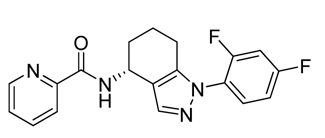
ML206
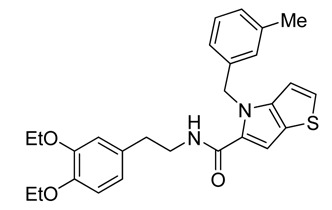
ML219
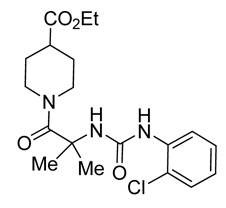
ML220
| CID/ML# | Target Name | IC50/EC50 (nM) [SID, AID] | Anti-target Name(s) | IC50/EC50 (μM) [SID, AID] | Fold Selective | Secondary Assay(s) Name: IC50/EC50 (nM) [SID, AID] |
|---|---|---|---|---|---|---|
| CID 42288156/ML206 | Cellular phenotype for lipid droplet storage | 8 nM [SID 99381576, AID 488913] | Cytotoxicity | 8.9 μM [SID 99381576, AID 488908] | >1000-fold | HeLa cell lipid droplet storage; HepG2 cell lipid droplet storage; AML12 cell lipid droplet storage; SID: 99381576 AID: 488900, 492960, 504434 |
| CID 46947845/ML219 | Cellular phenotype for lipid droplet storage | 2 nM [SID 99460848, AID 504432] | Cytotoxicity | 11.2 μM [SID 99460848, AID 504433] | >1000-fold | HepG2 cell lipid droplet storage; AML12 cell lipid droplet storage; SID: 99460848 AID: 492960, 504434 |
| CID 46947860/ML220 | Cellular phenotype for lipid droplet storage | 705 nM [SID 99460839, AID 504432] | Cytotoxicity | inactive [SID 99460839, AID 504433] | >1000-fold | HepG2 cell lipid droplet storage; AML12 cell lipid droplet storage; SID: 99460839 AID: 492960, 504434 |
Recommendations for Scientific Use of the Probe
Lipid droplets (LDs) are the universal lipid storage organelles. Lipids remobilized from LDs are used both for energy production and anabolic reactions, such as membrane biosynthesis. Nearly all mammalian components of adipocyte lipolysis are conserved in the fruit fly, Drosophila melanogaster, and the LDs of both species are coated with a similar set of proteins.1–9 These facts, combined with the rich genetic toolbox available for Drosophila melanogaster, positions the fruit fly as a prime in vivo model to study the mechanisms and pathways involved in lipid droplet biogenesis, regulation and breakdown. In this Drosophila melanogaster cell-based screen, the probes ML206, ML219 and ML220 show a potent LD reduction phenotype, with EC50s of 8 nM, 2 nM, and 705 nM, respectively. When looking at translation of this phenotype into mammalian cell lines, ML206 showed no significant activity in HepG2-, HeLa- or AML12-cells with <50% efficacy when compared to Triacsin C. Moderate translation into mammalian cells was seen for ML219, which gave an EC50 of 11 nM with 46% efficacy with AML12-cells. The third probe, ML220, also showed moderate translation in the AML12 cell line, reducing LD’s with an EC50 of 1.25 μM with >57% efficacy. With all three compounds showing extremely potent LD phenotypes in S3-cell based assays and modest translation into AML12-cells for ML219 and ML220, the utility of these compounds lies predominantly in Drosophila melanogaster studies, but they do show promise for use in mammalian systems. The target and mechanism of action for these probes in Drosophila melanogaster cells are not currently known, so studies using affinity chromatography, photoaffinity labeling and RNAi screening will hopefully elucidate their targets and provide insight into underlying mechanisms of LD biogenesis, maintenance and/or breakdown in mammalian systems.
2. Materials and Methods
General Methods for Chemistry. All air or moisture sensitive reactions were performed under positive pressure of nitrogen with oven-dried glassware. Anhydrous solvents such as dichloromethane, N,N-dimethylformamide (DMF), acetonitrile, methanol and triethylamine were obtained by purchasing from Sigma-Aldrich. Preparative purification was performed on a Waters semi-preparative HPLC. The column used was a Phenomenex Luna C18 (5 micron, 30 × 75 mm) at a flow rate of 45 mL/min. The mobile phase consisted of acetonitrile and water (each containing 0.1% trifluoroacetic acid). A gradient of 10% to 50% acetonitrile over 8 minutes was used during the purification. Fraction collection was triggered by UV detection (220 nM). Analytical analysis was performed on an Agilent LC/MS (Agilent Technologies, Santa Clara, CA). Method 1: A 7 minute gradient of 4% to 100% Acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with an 8 minute run time at a flow rate of 1 mL/min. A Phenomenex Luna C18 column (3 micron, 3 × 75 mm) was used at a temperature of 50°C. Method 2: A 3 minute gradient of 4% to 100% Acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with a 4.5 minute run time at a flow rate of 1 mL/min. A Phenomenex Gemini Phenyl column (3 micron, 3 × 100 mm) was used at a temperature of 50 °C. Purity determination was performed using an Agilent Diode Array Detector on both Method 1 and Method 2. Mass determination was performed using an Agilent 6130 mass spectrometer with electrospray ionization in the positive mode. 1H NMR spectra were recorded on Varian 400 MHz spectrometers. Chemical Shifts are reported in ppm with tetramethylsilane (TMS) as internal standard (0 ppm) for CDCl3 solutions or undeuterated solvent (DMSO-H6 at 2.49 ppm) for DMSO-d6 solutions. All of the analogs for assay have purity greater than 95% based on both analytical methods. High resolution mass spectrometry was recorded on an Agilent 6210 Time-of-Flight LC/MS system. Confirmation of molecular formula was accomplished using electrospray ionization in the positive mode with the Agilent Masshunter software (version B.02).
2.1. Assays
Table 1Screens Deposited in PubChem
| AID | Type | Target | Conc. Range | Samples Tested |
|---|---|---|---|---|
| 2685 | Primary qHTS | Lipid Storage in S3 cell | 46.2 μM – 3 nM | 321194 |
| 1569 | Confirmatory | Lipid Storage in S3 cell | 46.2 μM – 0.3 nM | 749 |
| 488913 | Confirmatory (for probe SAR) | Lipid Storage in S3 cell | 46.2 μM – 0.3 nM | 153 |
| 504432 | Confirmatory (for probe SAR) | Lipid Storage in S3 cell | 46.2 μM – 0.3 nM | 37 |
| 488908 | Anti-target (for probe SAR) | Cytotoxicity | 46.2 μM – 0.3 nM | 151 |
| 504433 | Anti-target (for probe SAR) | Cytotoxicity | 46.2 μM – 0.3 nM | 37 |
| 488900 | Secondary (for probe SAR) | Lipid Storage in Hela cell | 46.2 μM – 0.3 nM | 56 |
| 492960 | Secondary (for probe SAR) | Lipid Storage in HepG2 cell | 46.2 μM – 0.3 nM | 112 |
| 504434 | Secondary (for probe SAR) | Lipid Storage in AML12 cell | 46.2 μM – 0.3 nM | 132 |
| 1623 | Summary | Lipid Storage | NA | NA |
Primary qHTS assay for lipid storage modulators [AID:2685]
Assay details and protocol. The qHTS was performed with embryonic Drosophila S3 cells (Bloomington Drosophila Stock Center [DGRC]), that showed excellent oleic acid feeding characteristics and good performance during automated liquid handling in 1,536-well format (Table 2). We dispensed 4 μl of cells at 1.25 × 106 cells/ml into LoBase Aurora COC 1,536-well plates (black walled, clear bottom) with a bottle-valve solenoid-based dispenser (Kalypsys) to obtain 5,000 cells/well. A total of 23 nl of compound solution of different concentrations was transferred to the assay plates using a Kalypsys pin tool equipped with a 1,536-pin array containing 10-nl slotted pins (FP1S10, 0.457-mm diameter, 50.8 mm long; V&P Scientific). One microliter of oleic acid (400 μM) was added, and the plate was lidded with stainless steel rubber gasket-lined lids containing pinholes. After 18–24 h incubation at 24 °C and 95% humidity, 4 μl of BODIPY 493/503 (Molecular Probes) was added to the wells to stain lipid droplets, and the Cell Tracker Red CMTPX dye (Molecular Probes) was added to enumerate cell number. Fluorescence was detected by excitation of the fluorophores with a 488 nm laser on an Acumen Explorer (TTP Lab Tech). The total intensity in channel 1 (500–530 nm) reflected lipid droplet accumulation. Cells were detected using channel 3 (575–640 nm) with 5 μm width and 100 μm depth filters. The ratio of the total intensity in PMT channel 1 over total intensity of channel 3 was also calculated. Percent activity was computed relative to an internal control (100% inhibited lipid droplet deposition due to the presence of 20 μM Triacsin C), which was added to 32 wells/plate. Example data for the assay is shown in Figure 1.
Table 2
Stepwise protocol used for the S3 cell 1536-well assay.
Confirmatory assay. [AID 1569, AID 488913 and AID 504432]
Secondary assays. Cytotoxicity was tested using CellTiter-Glo to measure ATP levels using a final compound concentration of 46.2 μM to 0.3 nM and a 24 hr time point [AID 488908 and AID 504433]. To test for the possibility that compounds found to be active in the lipid S3 assay remained active in mammalian cell lines, we employed HeLa [AID 488900], HepG2 [AID 492960] and AML12 cells [AID 504434] with an analogous assay protocol (Table 3).
Table 3
Stepwise Protocol Used for the Hela, HepG2 or AML12 cell 1536-Well Assay.
2.2. Probe Chemical Characterization
ML206
1H NMR (400 MHz, DMSO-d6); 8.60 (m. 2H), 8.08 (m. 1H), 8.00 (m, 1H), 7.51–7.73 (m, 3H), 7.25 (m, 1H), 5.14 (m, 1H), 2.38–2.56 (m, 2H), 1.70–2.01 (m, 4H). Method 1, retention time, 5.336 min; Method 2, retention time 3.267 min; HRMS:° m/z °(M+) = 354.1298 (Calculated for C19H16F2N4O = 354.1292). [α]D +82 (c=0.22, MeOH). Solubility (PBS, pH 7.4, 23 °C) = 20.4 μg/mL. Stability profile over 48 hrs (PBS, pH 7.4, 23°C) is show in Figure 2. Calculated properties for ML206 are shown in Table 4.
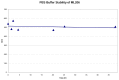
Figure 2
Stability of ML206 in PBS buffer (pH 7.4, 23°C) plotted as AUC vs. time for a 48 hr period using LC/MS Method 1 described in Section 2. No instability was observed, and >90% of compound remained at 48 hours.
Table 4
Probe properties.
Table 5Probe and analogs submitted to the MLSMR
ML219
1H NMR (400 MHz, DMSO-d6) δ ppm 8.29 (t, J=5.67 Hz, 1 H), 7.39 (d, J=5.48 Hz, 1 H), 7.08 – 7.15 (m, 2 H), 7.04 (s, 1 H), 6.96 – 7.02 (m, 2 H), 6.87 (d, J=7.43 Hz, 1 H), 6.75 – 6.83 (m, 2 H), 6.66 (dd, J=8.22, 1.96 Hz, 1 H), 5.71 (s, 2 H), 3.93 (qd, J=6.98, 4.70 Hz, 4 H), 3.34 – 3.43 (m, 2 H), 2.70 (t, J=7.24 Hz, 2 H), 2.19 (s, 3 H), 1.25 (dt, J=10.27, 6.90 Hz, 6 H). Method 1, retention time, 6.811 min; Method 2, retention time 3.787 min; HRMS:° m/z °(M+H+) = 462.1985 (Calculated for C27H30N2O3S = 462.1977). Solubility (PBS, pH 7.4, 23 °C) = 5.9 μg/mL. Stability profile over 48 hrs (PBS, pH 7.4, 23 °C) is shown in Figure 3.
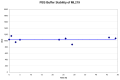
Figure 3
Stability of ML219 in PBS buffer (pH 7.4, 23°C) plotted as AUC vs. time for a 48 hr period, using LC/MS Method 1 described in Section 2. No instability was observed, and >90% of compound remained at 48 hours.
Table 6Probe and analogs submitted to the MLSMR
| NCGC ID | Probe/Analog | MLS ID | SID |
|---|---|---|---|
| NCGC00241426-02 | Probe (ML219) | MLS003221395 | 99460848 |
| NCGC00238551-02 | Analog | MLS003224199 | 143471935 |
| NCGC00241426-02 | Analog | MLS003221395 | 160779721 |
| NCGC00241438-01 | Analog | MLS003221396 | 99460862 |
| NCGC00241435-01 | Analog | MLS003221397 | 99460860 |
| NCGC00241443-01 | Analog | MLS003221398 | 99460869 |
| NCGC00241420-01 | Analog | MLS003221399 | 99460871 |
| NCGC00241419-01 | Analog | MLS003221400 | 99460865 |
ML220
1H NMR (400 MHz, DMSO-d6) δ ppm 8.03 (dd, J=8.41, 1.56 Hz, 1 H), 7.94 (s, 1 H), 7.42 (s, 1 H), 7.35 (dd, J=8.02, 1.37 Hz, 1 H), 7.18 (ddd, J=8.41, 7.24, 1.57 Hz, 1 H), 6.85 – 6.96 (m, 1 H), 4.28 (dt, J=13.16, 3.30 Hz, 2 H), 3.96 (q, J=7.17 Hz, 2 H), 2.76 – 2.99 (m, 2 H), 2.43 – 2.55 (m, 1 H), 1.73 (dd, J=13.11, 2.93 Hz, 2 H), 1.31 – 1.45 (m, 8 H), 1.06 (t, J=7.14 Hz, 3 H). Method 1, retention time, 4.995 min; Method 2, retention time 3.281 min; HRMS:° m/z °(M+) = 395.1623 (Calculated for C19H26ClN3O4 = 395.1612). Solubility (PBS, pH 7.4, 23 °C) = >75.0 μg/mL. Stability profile over 48 hrs (PBS, pH 7.4, 23 °C) is shown in Figure 4.
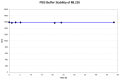
Figure 4
Stability of ML220 in PBS buffer (pH 7.4, 23°C) plotted as AUC vs. time for a 48 hr period, using LC/MS Method 1 described in Section 2. No instability was observed, and >90% of compound remained at 48 hours.
Table 7Probe and analogs submitted to the MLSMR
| NCGC ID | Probe/Analog | MLS ID | SID |
|---|---|---|---|
| NCGC00241450-01 | Probe (ML220) | MLS003390985 | 99460839 |
| NCGC00242556-01 | Analog | MLS003370615 | 110167689 |
| NCGC00242559-01 | Analog | MLS003370616 | 110167692 |
| NCGC00242560-01 | Analog | MLS003370617 | 110167693 |
| NCGC00242553-01 | Analog | MLS003370618 | 110167686 |
| NCGC00242554-01 | Analog | MLS003370619 | 110167687 |
| NCGC00242557-01 | Analog | MLS003370620 | 110167690 |
2.3. Probe Preparation
Synthesis of ML206
Scheme 1, Steps 1-3 were adapted directly from Guo et al.14
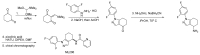
Scheme 1
Synthesis of enantiomerically pure ML206.
Scheme 1, Step 4. 1-(2,4-difluorophenyl)-4,5,6,7-tetrahydro-1H-indazol-4-amine (0.1 g, 0.401 mmol) and picolinic acid (0.049 g, 0.401 mmol) were dissolved in DMF (2 ml) and DIPEA (0.091 ml, 0.522 mmol) was added, followed by HATU (0.183 g, 0.481 mmol); the reaction was stirred at RT for 2 hrs, then purified by directly injecting to a Waters® reverse phase purification system.
Scheme 1, Step 5. Chiral chromotography was performed on a Chiralpac IB column (2 cm × 25 cm, 20 micron) using EtOH/Hexane (60/40, v/v) as the eluant at a flow rate of 4.0 mL/min, with a 30 min run time to give enantiomerically pure ML206.
Synthesis for determination of absolute configuration of ML206
Scheme 4, Step 6. NaOH (0.762 ml, 0.762 mmol), then (Boc)2O (0.195 ml, 0.838 mmol) were added to the solution of racemic 1-(2,4-difluorophenyl)-4,5,6,7-tetrahydro-1H-indazol-4-amine (0.19 g, 0.762 mmol) in DCM (8 ml). The mixture was stirred at RT for 3hrs. The organic layer was washed with H2O twice, dried over Na2SO4, and concentrated. The crude product was submitted for chiral separation.
Scheme 4, Step 7. Chiral chromatography was performed on a Chiralpac OJ column (20 × 50 cm, 20 micron) with MeOH as the eluant, a flow rate of 40 mL/min and a 40 min run time.
Scheme 3, Step 8. TFA (0.2 ml, 2.60 mmol) was added to the solution of the optically pure tert-butyl 1-(2,4-difluorophenyl)-4,5,6,7-tetrahydro-1H-indazol-4-ylcarbamate (17 mg, 0.049 mmol) in DCM (1 ml). The mixture was stirred at RT overnight. The solvent was then removed in vacuo and placed on a high vacuum pump (~1 torr) overnight. The crude product was used in the next reactions without purification.
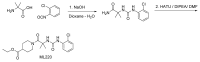
Scheme 3
Synthesis of ML220.
Scheme 3, Step 9-11.
The enantiomerically pure amine was coupled with either picolinic acid, R-Mosher acid or S-Mosher acid using the amide coupling described for step 4 scheme 1. All products were purified using a Waters® reverse phase purification system.
Synthesis of ML219
Scheme 2, Step 1. HATU (0.751 g, 1.974 mmol) and DIPEA (0.94 ml, 5.38 mmol) were added to a solution of 4H-thieno[3,2-b]pyrrole-5-carboxylic acid (0.3 g, 1.794 mmol) in DMF (25 ml). The solution was stirred at room temperature for 30 min, and then 2-(3,4-diethoxyphenyl)ethanamine (0.413 g, 1.974 mmol) was added. The reaction was stirred at room temperature overnight. Water (40 ml) was added to the mixture. The solid was filtered and washed with water, and dried. The crude product was used in the next reaction without further purification.
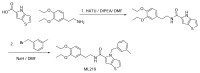
Scheme 2
Synthesis of ML219.
Scheme 2, Step 2. Sodium hydride (3.68 mg of 60 wt% dispersion in mineral oil, 0.092 mmol) was added to the solution of N-(3,4-diethoxyphenethyl)-4H-thieno[3,2-b]pyrrole-5-carboxamide (30 mg, 0.084 mmol) in DMF (1.5 ml). The mixture was stirred at room temperature for 15 min. 1-(bromomethyl)-3-methylbenzene (0.014 ml, 0.1 mmol) was added to this mixture. The reaction mixture was stirred at room temperature overnight. The DMF solution was purified by directly injecting to a Waters® reverse phase purification system to give ML219 as a TFA salt.
Synthesis of ML220
Scheme 3, Step 1. A solution of NaOH (0.028 g, 0.698 mmol) in Water (1 ml) was added to a suspension of 1-aminocyclohexanecarboxylic acid (0.1 g, 0.698 mmol) in 1,4-dioxane (3 ml) at RT. The mixture was stirred at RT for 20 min (clear solution). A solution of 2,4-dichloro-1-isocyanatobenzene (0.131 g, 0.698 mmol) in dioxane (2 ml) was added dropwise to the reaction mixture at RT. The resulting solution was stirred at RT for 1 hr, and then evaporated to dryness under reduced pressure to give the title compound.
Scheme 3, Step 2. HATU (81 mg, 0.214 mmol), then DIPEA (0.102 ml, 0.584 mmol) were added to a solution of 2-(3-(2-chlorophenyl)ureido)-2-methylpropanoic acid (50 mg, 0.195 mmol) in DMF (1.5 ml). The solution was stirred at RT for 30 min, and then ethyl piperidine-4-carboxylate (0.033 ml, 0.214 mmol) was added. The reaction was stirred at RT for 3 hrs. The DMF solution was purified by directly injecting to a Waters® reverse phase purification system to give ML220 as a TFA salt.
3. Results
3.1. Dose Response Curves for Probe
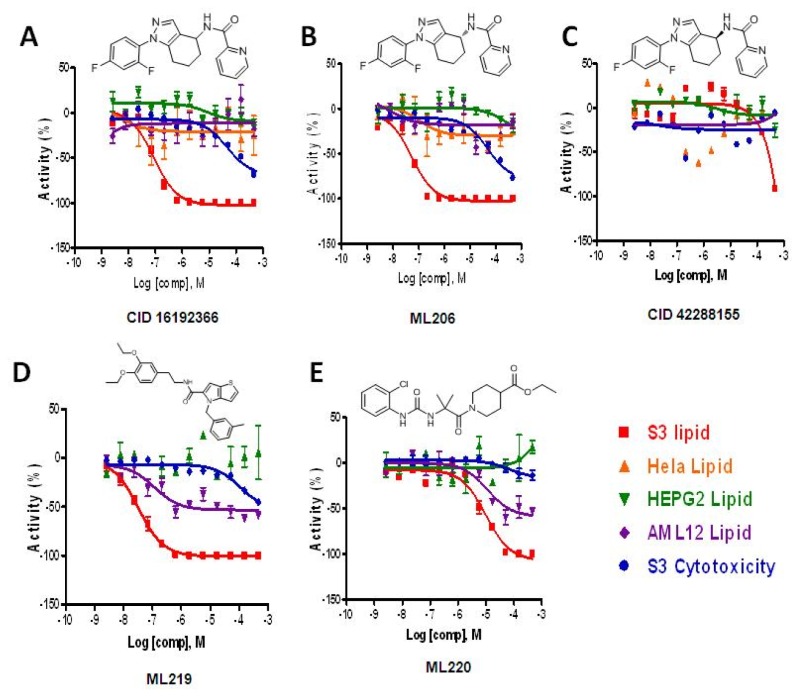
Figure 5Potency confirmation
Confirmatory data for the probe compounds in S3, Hela, HepG2 and AML12 cells, along with cytotoxicity determined in S3 cells. The lipid reduction % Activity plotted is from the lipid data. A. The racemic compound CID16192366. B. The enantiomerically pure compound where the absolute configuration was confirmed as (R) for ML206. C. The enantiomerically pure compound where the absolute configuration was confirmed as (S) for CID42288155. D. The probe compound ML219 which also showed weak activity in AML12 cells. E. The probe compound ML220 which also showed weak activity in AML12 cells.
The enantiomerically pure compounds have also been transferred to the assay provider’s lab, and Dr. Beller has confirmed activity in S3 cells using the same lipid-specific BODIPY dye, but with wide-field microscope-based analysis (Figure 6). These results support high and low potency for the (R) and (S) enantiomers respectively, as well as low cytotoxicity in S3 cells.
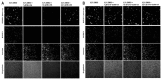
Figure 6
BODIPY and Hoechst staining of S3 cells treated with compounds. Data was collected at the Max Planck Institute for Biophysical Chemistry, Department of Molecular Developmental Biology.
3.2. Cellular Activity
Primary and all follow-up assays were performed in Drosophila melanogaster cells (S3), HeLa, HepG2 and AML12 cells, with activity seen in varying levels in 3 of these cell lines (S3, HepG2 and AML12). Therefore, the compounds are deemed to have sufficient cell permeability/activity. No significant cytotoxicity (as compared to EC50 values) was seen for these probes (and the majority of their analogs) in any of the 4 cell lines. ML206, ML219 and ML220 showed >1000-fold cytotoxicity windows in S3-cells, with only minimal cytotoxicity at 46 μM (Figure 6).
3.3. Profiling Assays
Profiling assays have not been done with these probes, as they show minimal activity in mammalian cell-lines, and thus such profiles are not deemed valuable at this point.
4. Discussion
4.1. Comparison to Existing Art and How the New Probe is an Improvement
We have previously reported that inhibition of COPI function by Exo1 (ML084) results in lipid overstorage, but we had not discovered inhibitors of LD accumulation at that time.16 Few substances are available as reagents for in vitro lipid storage research (i.e. Triacsin C15, Exo1 (ML084)16,17, Isoproterenol and other β-adrenergic agonists18 and natural products, such as Magnolol19). While these compounds are useful, the biogenesis, maintenance, breakdown, signaling pathways and mechanisms associated with LDs are not fully understood, and the discovery of novel and potent LD storage modulators is still highly sought. Particularly, the identification of compounds targeting new proteins is desirable to help shed light on specific aspects of these pathways. The previous probes discovered (ML206 and ML207 (CID 46916320)) are extremely potent lipid storage inhibitors in Drosophila melanogaster, and the new additions (ML219 and ML220) show similar potent phenotypes in the S3-cell line with improved translation into the mammalian cell line. This renders these new probes as considerable improvements over the existing art. Currently, there is an ongoing effort to discover the target(s) of all three probes using photoaffinity labeling, affinity chromatography and RNAi screening experiments.
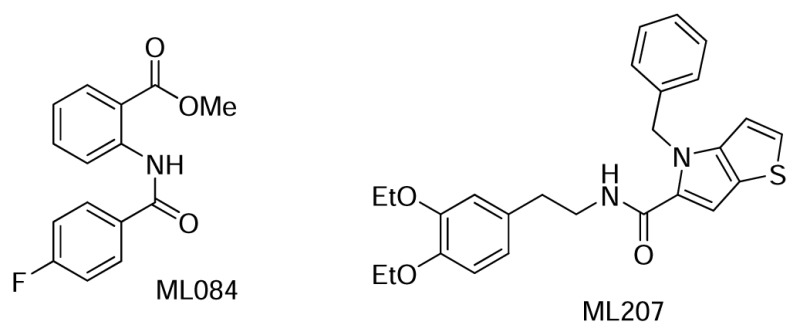
Figure 7Previous ML probes that modulate lipid storage in S3 cells
5. References
- 1.
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–75. [PubMed: 10399915]
- 2.
- Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–6. [PubMed: 12676093]
- 3.
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–30. [PubMed: 16054079]
- 4.
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. [PMC free article: PMC1865564] [PubMed: 17488184]
- 5.
- Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–81. [PubMed: 14550535]
- 6.
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–65. [PubMed: 17608402]
- 7.
- Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–94. [PubMed: 16543254]
- 8.
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–95. [PubMed: 16979555]
- 9.
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–92. [PubMed: 14597625]
- 10.
- Brasaemle DL. J Lipid Res. 2007;48:2547–2559. [PubMed: 17878492]
- 11.
- Gieselmann V. Biochimica et Biophysica Acta. 1995;1270:103–136. [PubMed: 7727535]
- 12.
- Winchester B, Vellodi A, Young E. Biochem Soc Trans. 2000;28:150–154. [PubMed: 10816117]
- 13.
- Chien S, Reiter LT, Bier E, Gribskov M. Nucleic Acids Res. 2002;30:149–151. [PMC free article: PMC99119] [PubMed: 11752278]
- 14.
- Guo S, Song Y, Huang Q, Yuan H, Wan B, Wang Y, He R, Beconi MG, Franzblau SG, Kozikowski AP. J Med Chem. 2010;53:649–659. [PubMed: 20000470]
- 15.
- Igal RA, Wang P, Coleman RA. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem J. 1997;324(Pt 2):529–34. [PMC free article: PMC1218462] [PubMed: 9182714]
- 16.
- Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:2530–2549. [PMC free article: PMC2586367] [PubMed: 19067489]
- 17.
- Feng Y, Yu S, Lasell TK, Jadhav AP, Macia E, Chardin P, Melancon P, Roth M, Mitchison T, Kirchhausen T. Exo1: a new chemical inhibitor of the exocytic pathway. Proc Natl Acad Sci U S A. 2003;100:6469–74. [PMC free article: PMC164470] [PubMed: 12738886]
- 18.
- Wright EE, Simpson ER. Inhibition of the lipolytic action of beta-adrenergic agonists in human adipocytes by alpha-adrenergic agonists. J Lipid Res. 1981;22:1265–70. [PubMed: 6119348]
- 19.
- Chen JS, Chen YL, Greenberg AS, Chen YJ, Wang SM. Magnolol stimulates lipolysis in lipid-laden RAW 264.7 macrophages. J Cell Biochem. 2005;94:1028–37. [PubMed: 15597343]
- PMCPubMed Central citations
- PubChem BioAssay for Chemical ProbePubChem BioAssay records reporting screening data for the development of the chemical probe(s) described in this book chapter
- PubChem SubstanceRelated PubChem Substances
- PubMedLinks to PubMed
- Review ML360, A Potent Inhibitor of Lipid Droplet Formation.[Probe Reports from the NIH Mol...]Review ML360, A Potent Inhibitor of Lipid Droplet Formation.Liu L, Zhang YQ, Tschapalda K, Li Z, Shen M, Beller M, Boxer MB. Probe Reports from the NIH Molecular Libraries Program. 2010
- The lipid-droplet proteome reveals that droplets are a protein-storage depot.[Curr Biol. 2006]The lipid-droplet proteome reveals that droplets are a protein-storage depot.Cermelli S, Guo Y, Gross SP, Welte MA. Curr Biol. 2006 Sep 19; 16(18):1783-95.
- Review The contribution of the Drosophila model to lipid droplet research.[Prog Lipid Res. 2011]Review The contribution of the Drosophila model to lipid droplet research.Kühnlein RP. Prog Lipid Res. 2011 Oct; 50(4):348-56. Epub 2011 May 17.
- Review Lipid droplets in lipogenesis and lipolysis.[Endocrinology. 2008]Review Lipid droplets in lipogenesis and lipolysis.Ducharme NA, Bickel PE. Endocrinology. 2008 Mar; 149(3):942-9. Epub 2008 Jan 17.
- Review Potent inhibitors of lipid droplet formation.[Probe Reports from the NIH Mol...]Review Potent inhibitors of lipid droplet formation.Zou J, Ganji S, Pass I, Ardecky R, Peddibhotla M, Loribelle M, Heynen-Genel S, Sauer M, Pass I, Vasile S, et al. Probe Reports from the NIH Molecular Libraries Program. 2010
- Modulators of Lipid Storage - Probe Reports from the NIH Molecular Libraries Pro...Modulators of Lipid Storage - Probe Reports from the NIH Molecular Libraries Program
Your browsing activity is empty.
Activity recording is turned off.
See more...