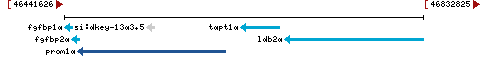- Exon count:
- 31
| Annotation release |
Status |
Assembly |
Chr
|
Location |
|
106
|
current |
GRCz11 (GCF_000002035.6) |
14 |
NC_007125.7 (46456088..46616591, complement) |
| 105 |
previous assembly |
GRCz10 (GCF_000002035.5) |
14 |
NC_007125.6 (47469007..47629506, complement) |
Chromosome 14 - NC_007125.7