NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Institute of Medicine (US) Forum on Emerging Infections; Davis JR, Lederberg J, editors. Public Health Systems and Emerging Infections: Assessing the Capabilities of the Public and Private Sectors: Workshop Summary. Washington (DC): National Academies Press (US); 2000.

Public Health Systems and Emerging Infections: Assessing the Capabilities of the Public and Private Sectors: Workshop Summary.
Show detailsOVERVIEW
Infectious disease surveillance is the first link in the response to emerging infections. The United States spent $74.5 million in 1992 for all infectious disease surveillance activities (Osterholm et al., 1993). Although the total amount of surveillance has increased in recent years, there have been dramatic cutbacks in surveillance for human immunodeficiency virus (HIV) infection, tuberculosis, and vaccine-preventable diseases (Michael Osterholm, state epidemiologist and chief, Minnesota Department of Health, personal communication, November 1998).
Infectious disease surveillance concurrently involves the health care delivery system, the public health laboratory, and epidemiologists. Each of these sectors contributes to the four basic components of surveillance, which are (1) collection, (2) analysis, (3) dissemination, and (4) response. Collection and analysis can be conducted at the local, state, federal, or international level by public agencies as well as by private industry. Dissemination and response are specific public health activities. Thus, disease surveillance is the ongoing, systematic collection and analysis, interpretation, and feedback of outcome-specific data. As such, surveillance may monitor cases of disease reported by clinicians or identified in laboratories, or it may monitor changes in practice or other behaviors of public health importance.
Relevant activities at the federal level include assessment of surveillance programs, funding of state activities, provision of services, and nationwide disease surveillance. At the state level, health departments establish the systems by which infection and disease are reported, data are gathered, and prevention is initiated. State and local health departments also play a vital role in educating physicians about the need to report and gather appropriate data. In the private sector, pharmaceutical companies and organizations that provide diagnostic services play a critical role in conducting large-scale surveillance for drug and vaccine development and in providing clinical data for retrospective and prospective studies.
Surveillance information is derived from many sources, and approaches to surveillance may vary depending on the kind of information that is needed and the resources that are available. Surveillance information is used in a variety of ways: to identify cases for investigation, to estimate magnitude of disease, to detect outbreaks, to evaluate response and prevention measures, to monitor changes in infectious agents, to facilitate research, and to measure the impacts of changes in health care practices.
Although there are common uses of surveillance data at the local, state, and federal levels, emphases vary. For example, individual case investigation is critical at the local and state levels but less critical at the federal level, unless a multijurisdictional disease outbreak occurs. Evaluation of larger-scale prevention and control measures—for example, the impact of new vaccines—is a high priority at the federal level. A national surveillance plan should take into account this diversity in the uses of data, approaches, and emphases at different government levels.
GAO REPORT ON PUBLIC HEALTH SURVEILLANCE OF EMERGING INFECTIOUS DISEASES
Authors
Marsha Lillie-Blanton, Ph.D., Associate Director and Helene Toiv, M.P.A., Assistant Director.Affiliations
A number of changes in the health care system—such as consolidations and managed care—combined with recent outbreaks of infectious diseases thought to be under control have led policy makers to evaluate, among other things, the functioning of public health and clinical laboratories. One such assessment was conducted by the U.S. General Accounting Office (GAO), a research agency of the U.S. Congress.
The GAO focused its effort on four tasks: (1) to describe the roles of different categories of private and public laboratories; (2) to gather information on the extent to which state surveillance programs and state public health laboratories contribute to the surveillance of six specific diseases;1 (3) to define the problems faced by public health officials, particularly at the state level, in gathering and using laboratory-generated information; and (4) to report on the role of the Centers for Disease Control and Prevention (CDC), particularly the kind of assistance it provides to the states. To conduct the assessment, GAO surveyed all state public health laboratory directors and all state epidemiologists and conducted case studies in Oregon, Kentucky, and New York.
GAO’s survey gathered information about the kinds of tests that the state public health laboratories performed in connection with the selected diseases, whether the state epidemiology program includes the six diseases in its surveillance program, and reporting requirements. Data were also collected on resources, both financial and human, and electronic communications. State officials were also asked about the assistance they received from CDC (see Box 3-1).2 Summary findings from the survey are found in Appendix C.
EMERGING INFECTIONS PROGRAM COOPERATIVE AGREEMENT
Authors
Robert Pinner, M.D.Affiliations
Following publication of the 1992 Institute of Medicine (IOM) report Emerging Infections: Microbial Threats to Health in the United States (IOM, 1992), CDC developed a plan to address emerging infections. It was released in April 1994, and was recently revised and updated. Since then, progress has been made in implementing this plan. The plan has the following four goals: (1) surveillance and response, (2) applied research, (3) infrastructure and training, and (4) prevention and control. In addition, among other activities, CDC sponsors the Emerging Infectious Diseases Journal, the CDC Association of Public Health Laboratory Directors, and public health laboratory fellowships, and provides support for World Health Organization collaborating centers.
CDC’s emerging infectious disease surveillance strategy makes an effort to take into account the diversity of needs and approaches through three complementary cooperative agreement programs: (1) the Epidemiology and Laboratory Capacity (ELC) program, (2) the Emerging Infections Program (EIP) network, and (3) provider-based sentinel networks. (See Figure 3-1 for a map of program locations; for more information, see the CDC website: www.cdc.gov.)
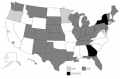
FIGURE 3-1
Map of Emerging Infections Programs (EIP) and Epidemiology and Epidemiology and Laboratory Capacity (ELC) Cooperative Agreements. SOURCE: CDC, 1998.
The purpose of the ELC program is to assist state and large local public health agencies in strengthening their basic epidemiological and laboratory capacities. It currently covers 30 jurisdictions, with full implementation of ELCs expected by the year 2002. (Essentially there would be ELCs in 63 jurisdictions, including states, large cities, and territories.) Health departments use these funds in a variety of ways, although the amount of total resources available is small relative to the needs. The ELC program has highlighted the challenge to define more clearly the required core laboratory and epidemiological capacities for the local, state, regional, and national levels and for various program areas, such as infectious diseases, foodborne diseases, and bioterrorism. A variety of activities are pursued in ELC program cooperative agreements, including enhanced information exchanges, training, education, and laboratory capacity, particularly including Pulsed Field Gel Electrophoresis (PFGE). Some states have been emphasizing collaborations with local health departments.
Eight EIP network sites distribute their activities among a variety of emerging infections program foci, including activities that address foodborne diseases (FoodNet); a family of project activities that involve invasive bacterial diseases (Active Bacterial Core surveillance, or ABCs); and surveillance for unexplained deaths and critical illnesses. Other activities include programs on judicious antibiotic use, surveillance for hepatitis, and enhancing the rate of immunization against pneumococcal infection. Full implementation of EIPs (in 10 jurisdictions total) is expected by the year 2002.
Population-based surveillance information plays a critical role across the EIP network. For example, all the sites perform active laboratory-based surveillance for invasive pneumococcal disease. Cases are defined by the isolation of Streptococcus pneumoniae from normally sterile sites, usually blood or cerebrospinal fluid. Such cases are sought actively by regular contact with the laboratories. Efforts are made to ensure that the data are comparable across sites and that there is essentially complete case ascertainment. This approach requires considerable effort and coordination among the sites and at CDC.
During 1995 and 1996 the incidence of invasive pneumococcal disease varied substantially across the sites, from 14.2/100,000 population in Toronto to more than 30/100,000 population in California and Georgia (see Figure 3-2). A convenience sample of pneumococcal isolates from assorted laboratories could not be used to characterize incidence in this way or demonstrate with confidence geographic variations in incidence. Rates of invasive pneumococcal disease vary considerably by age and race, which, along with the geographic distribution of underlying diseases like HIV infection that predispose individuals to pneumococcal disease, account for the geographic variations in the incidence of invasive pneumococcal disease.
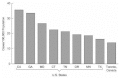
FIGURE 3-2
Incidence of Streptococcus pneumoniae disease, by state, July 1995 to June 1996. SOURCE: Cetron et al., 1997.
Active surveillance entails the collection of isolates, as well as case reports. Through such means of active surveillance, isolates have been tested at a common laboratory by reliable methods to measure penicillin susceptibility. The proportion of penicillin-resistant and -intermediate resistant isolates varies by site (see Figure 3-3). The distribution of penicillin resistance and intermediate resistance also varies by age, with the highest proportion of resistant and intermediate isolates found in young children. In some areas the distribution varies by race with whites having a higher proportion of infection with resistant isolates than blacks (though blacks have a higher overall incidence of pneumococcal disease).
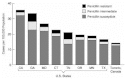
FIGURE 3-3
Incidence of invasive Streptococcus pneumoniae disease by geographic area and penicillin susceptibility. SOURCE: Cetron et al., 1997.
Another example of population-based surveillance in the EIPs is a set of activities called FoodNet. FoodNet is designed to determine and monitor the burden of foodborne diseases and improve understanding of the proportion of foodborne diseases attributable to various pathogens. Active laboratory-based surveillance along with laboratory, physician, and population surveys are used together to obtain estimates of the foodborne disease burden. For example, Campylobacter is the most common of the bacterial foodborne pathogens in FoodNet in this surveillance, followed by Salmonella and Shigella.
Population-based surveillance reveals seasonal, geographic, and demographic variations in foodborne diseases. For example, in the San Francisco Bay area, where the rates of foodborne illnesses caused by Campylobacter are the highest, rates of illness are much higher among Latino and Asian children than among other groups. This has prompted a case-control study of the risk factors for foodborne Campylobacter infections3 in Latino and Chinese-American children in the California EIP.
Surveillance for Campylobacter infections has also enabled a focused look at quinolone resistance in Minnesota. The proportion of C. jejuni isolates resistant to nalidixic acid increased over time. These infections occur in persons with a history of foreign travel, reflecting foreign exposure to resistant organisms. Increasingly, however, resistance is being observed among domestically acquired cases. This provides another example of how population-based surveillance data are enhanced by a focused look at an important public health phenomenon.
The highest incidence of Escherichia coli O157:H7 infection has also occurred in Minnesota. However, an EIP network-wide survey on physician knowledge of laboratory practices showed that physicians in Minnesota were more knowledgeable about whether their laboratories routinely cultured clinical specimens for E. coli O157:H7. They were also more likely than physicians at other sites to have actually obtained a sample for E. coli O157:H7 culture from their last patient with bloody diarrhea, suggesting that at least some of this apparent difference in incidence may be attributable to variations in practice.
Recent changes in the epidemiology of bacterial meningitis in the United States have resulted from the decline to very low levels of Haemophilus influenzae type b (Hib) due to the licensing and use of effective conjugate vaccines. At the same time, however, Group B streptococcal infections have emerged as an important cause of neonatal meningitis and sepsis in the United States; they are responsible for an estimated 7,500 cases of sepsis4 and meningitis5 annually among newborns with direct costs of $300 million. Accordingly, the EIP network has been using active, population-based surveillance to track the incidence of this disease, and also to gauge the uptake and impact of Group B streptococcal infection prevention policies.
In 1996, CDC, together with the Academy of Pediatrics and the College of Obstetricians and Gynecologists, developed consensus guidelines for the prevention of Group B streptococcal disease. These guidelines included late prenatal screening to identify Group B streptococcus carriers, intrapartum prophylaxis of preterm deliveries and carriers, and empiric prophylaxis based on risk factors for Group B streptococcus disease at labor. Surveillance data later showed a decline in the incidence of neonatal Group B streptococcal infection, but only in the incidence of disease with onset early in life. This is essentially what would be predicted if preventive policies were in place (see Figure 3-4).
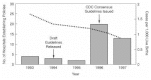
FIGURE 3-4
Number of hospitals in California, Georgia, Maryland, and Tennessee establishing Group B streptococcus prevention policies and the incidence of early-onset Group B streptococcal disease by year, based on active surveillance. SOURCE: Adapted from Rosenstein (more...)
Additional data indicate that hospitals with no Group B streptococcal prevention policy had little or no decrease in the mean number of cases of Group B streptococcus infection between 1996 and 1997, but hospitals that adopted or revised a Group B streptococcus prevention policy in 1996 saw a significant decline in the number of early onset cases of Group B streptococcal infection (see Table 3-1).
TABLE 3-1
Early-Onset Group B Streptococcus (GBS) in Hospitals with and Without Prevention Policies in 1996.
In conclusion, the EIP Network has been a valuable component in the implementation of CDC’s plan to address emerging infections by emphasizing population-based active surveillance and collaboration among CDC and partners in the public health and academic communities.
LARGE NATIONAL COMMERCIAL LABORATORIES
Authors
Linda Miller, Ph.D.Affiliations
Private companies, such as SmithKline Beecham Pharmaceuticals, have an interest in antimicrobial agents and vaccines. To pursue these interests, antimicrobial susceptibility surveillance studies are conducted to determine the percentage of organisms that are susceptible to different antimicrobial agents.
Large commercial laboratories have several reasons to pursue surveillance: (1) quality assurance, (2) as a service to customers (e.g., for respiratory infections, data on susceptibility could affect empiric therapy) (3) compliance with federal regulatory agencies through the provision of data on reportable diseases, (4) as a service to current public health, and (5) product development.
In 1993, the FDA requested that pharmaceutical companies gather and submit in vitro susceptibility data for organisms that were listed on the product information label (package insert available for prescription drugs). As a result, pharmaceutical companies have an interest in gathering in vitro susceptibility data on organisms. In addition, the pharmaceutical industry conducts antimicrobial surveillance to demonstrate the activity of a drug in comparison with those of other agents.
Such surveillance studies usually include isolates from community-acquired or hospital-based infections. These studies are conducted to track rates of resistance, in part to ensure that the drug maintains its activity so that the organism can remain on the product labeling.
For drugs that are in development, surveillance is also critical as an adjunct to or in support of clinical trials. Some clinical studies are required to establish that a drug has efficacy against resistant pathogens, which is difficult if the rates of resistance are low. Moreover, because of the study design, a majority of isolates from the patients in the clinical study may not be highly resistant, requiring supplemental in vitro studies.
Traditional surveillance entails large-scale studies over a wide geographic region to collect isolates for susceptibility testing. Targeted surveillance involves searching for geographic areas where isolates are resistant. This type of surveillance is useful when developing novel antimicrobial agents. Molecular or genetic-based studies are also used for surveillance. Finally, existing databases, such as those created through routine susceptibility testing, provide another opportunity for surveillance.
The SmithKline Beecham Clinical Laboratories (now Quest Laboratories) are among the largest clinical laboratories in the United States, with more than 80,000 physicians, hospitals, and corporate clients and more than 100 million tests processed annually. One of its efforts is Project Beta-Alert, a Haemophilus study that aims to determine the percentage of beta-lactamase-producing Haemophilus influenzae strains among the H. influenzae isolates sent to the large commercial reference laboratory. The project includes isolates from most states. As part of the routine daily work, a technologist isolates the organism, identifies it as H. influenzae, and performs the beta-lactamase test. The data can be tracked to a zip code. Data from 1997, sorted by specimen source, patient age, month of collection, laboratory site, state, and zip code, suggest that for empiric therapy, 3 in 10 patients require treatment with a drug resistant to beta lactamase activity. Data collected from 1993 to 1997 for 44,691 Haemophilus isolates showed that 35 percent produced a beta-lactamase, with the highest percentage of isolates being from children under age 6. Another study conducted in 1997 and 1998 involved collection of 2,000 isolates each of Streptococcus pneumoniae and H. influenzae. Isolates were collected from SmithKline Beecham laboratory sites in all regions of the United States and were sent to a central laboratory for testing. For S. pneumoniae, the highest rate of penicillin resistance was found among isolates in samples from the south-central and southeast regions of the United States. Macrolide resistance was also highest among isolates in samples from the south central and southeast regions of the United States. The surveillance data also showed rates of penicillin resistance in S. pneumoniae by site of infection, with the ear and sinus isolates having the highest rates of penicillin resistance. Data on age distribution revealed that the vast majority of the drug-resistant S. pneumoniae and Haemophilus isolates were found in young children less than 2 years of age.
Examples of other large surveillance projects in the planning stage include a pneumococcal surveillance project, the development of a national database on gram-positive and gram-negative organisms, and surveillance for drug resistance among Mycobacterium tuberculosis strains. Studies that attempt to correlate antimicrobial agent use with antimicrobial agent resistance, a relationship that is difficult to establish, are also being considered.
Some companies provide in vitro data from an on-line surveillance network, which connects hospitals with company computers on a nightly basis. Data on all the organism identifications and the results of susceptibility testing are collected. However, because samples come from many states and states vary in their reporting practices, data collection can be arduous. In addition, a networked system between the private and public sectors requires a certain level of data standardization. This type of routine data collection is an invaluable resource in retrospective analyses for surveillance purposes.
Any large-scale private surveillance effort must guard against violation of patient privacy as well as breach of proprietary concerns. Because diseases cross borders, such commercial work must also contend with international issues. Moreover, large-scale surveillance is expensive and profit margins are slim for commercial clinical laboratories. Given these limitations, it will be increasingly important for the public and private sectors to arrange collaborative surveillance projects on matters of widespread public health consequence.
ROLE OF THE PUBLIC-SECTOR LABORATORY AT THE NATIONAL LEVEL
Authors
Joe McDade, Ph.D.Affiliations
In 1988, IOM defined public health as what society does collectively to ensure the conditions in which people can be healthy (IOM, 1988). The core functions of public health are assessment, policy development, and assurance. Activities performed by the public health sector include the following:
- population-based disease surveillance;
- epidemiological investigations;
- environmental assessments of food, water, and vectors;
- ensuring the quality of public-and private-sector laboratory testing;
- tracking incipient trends;
- conducting outbreak investigations;
- tracking the distribution and the migration of noteworthy pathogens; and
- monitoring the effectiveness of prevention programs.
CDC laboratories make a unique contribution to reference diagnostic or confirmatory testing. CDC also plays a pivotal role in gathering, collating, and analyzing data from multiple sources. For example, reports of deaths due to pneumonia and influenza come to CDC from many sources: state health departments, physicians, and a World Health Organization network comprising 110 different laboratories, in 83 different countries, that obtain isolates and use reagents and materials provided by CDC to determine the influenza virus type. CDC can further subtype isolates submitted to it by using molecular sequencing. All these efforts ensure relevance of the influenza vaccines produced every year.
Another example of a key role of a national laboratory, such as CDC’s National Center for Infectious Diseases, is in the characterization of various measles virus isolates to determine if prevention efforts have stopped the indigenous transmission of measles. To conduct this work, isolates were obtained throughout the world and were then sequenced for determination of their genotypes. It was found that certain measles viruses are peculiar to certain areas of the world and that transmission is indigenous in those countries. The data also showed that a measles vaccination program had been effective in interrupting transmission of the measles virus in the United States and that the existing vaccine offered protection against those isolates reported from outside the United States.
CDC and other laboratories also serve as international reference laboratories for determination of whether polio virus types 1, 2, and 3 are being transmitted and whether polio virus infections are caused by wild-type strains or breakthroughs of the attenuated vaccine strain. Such information is crucial in immunization efforts.
Despite the strengths of the public-sector national laboratories, there is a need for collaboration with private-sector facilities in the standardization of databases and the evaluation of reagents and techniques. Such collaborations will be particularly important because cost-containment efforts and changes in the ways in which health care is administered may compromise disease surveillance efforts. For example, under managed care, there is a disincentive to collect isolates and specimens for cultures on which laboratory surveillance is based. In certain instances, the commercial laboratory system has supported the state public health laboratories when high-volume testing has been needed for tests for rare and unusual diseases. In contrast, public health laboratories are more suited to performing specialized testing.
In the absence of a well-defined national laboratory system, more strategic planning is needed. This would entail, for example, defining the surveillance and information needs for specific diseases, the type of testing needed, the materials and specimens required, the roles of the public and private sectors, and the referral systems and core capabilities, as well as standardizing methods and databases.
ROLE OF THE PUBLIC-SECTOR LABORATORY AT THE STATE LEVEL
Authors
Mary Gilchrist, Ph.D.Affiliations
Most state public health laboratories were instituted in the early part of the 20th century, when they were nearly the sole source of expertise and training in public health and when a microscope might have been the primary tool available for diagnosis of a disease. By mid-century there was a decline in the rate of infectious diseases and a rise in local laboratory expertise, which led to more focus at the state level on diseases of traditional public health importance such as tuberculosis, syphilis, and rabies and on specialized reference bacteriology and virology.
By the 1970s another type of state public health facility, the combined or consolidated laboratory, emerged when the U.S Environmental Protection Agency was instituted and some public health laboratories expanded to include environmental testing. A third type of laboratory, the consolidated laboratory, performs still other services for the state in the areas of agriculture, forensics, and some newborn screening.
By the mid-1990s, with the inception of the emerging infections program, CDC’s new cooperative agreements with states created new opportunities and challenges for state laboratories. They began expanded activities such as typing of Mycobacterium tuberculosis, Escherichia coli O157, and Salmonella isolates and characterization of antibiotic resistance. New molecular techniques such as these have established critical new roles for public health laboratories.
Most public health laboratories have routine surveillance programs for infectious agents of significance in their regions. Outbreaks are then detected earlier and are defined more clearly.
An example of an active state surveillance program is Iowa’s influenza virus surveillance activities. At the state laboratory the influenza virus is isolated, identified, and typed before being sent to CDC as part of the World Health Organization surveillance system that tracks disease and predicts vaccine needs. The state laboratory automatically posts nightly on the Internet (http://www.uhl.uiowa.edu) all of the influenza virus isolates in its database. Thus, real-time data on the geographic incidence of various influenza virus types and the number of isolates that have been detected in each of the state’s regions throughout the winter season are available. In the future, the system will offer quicker detection of more viral agents. It will also provide hypertext links to information on vaccines and antiviral therapies. Thus, if a clinician wants to know whether the current vaccine is effective against a given circulating strain, a query to the website will provide that information.
The public health laboratory in Iowa also conducts vector-borne disease surveillance. For example, it monitors sentinel chicken flocks for the presence of antibodies to arboviruses and examines pools of mosquitoes for the presence of arboviruses, enabling cities and counties to control mosquitoes before encephalitis6 viruses cause a substantial risk to humans. Surveillance for the emergence of tick-borne infections is also conducted in Iowa. Deer hunters send in samples of deer blood on filter paper for studies of the distribution of antibodies against the Lyme disease spirochete and the agents of ehrlichiosis. Surveillance for vector-borne diseases has shown how these pathogens are emerging in Iowa along the Mississippi River through animals that are migrating up the river valleys. As a result, public health laboratories can alert health care practitioners about the prevalence of new infectious agents in Iowa.
In collaboration with the University of Iowa, the state Hygienic Laboratory conducts an antibiotic resistance surveillance program, looking for organisms that cause invasive diseases: enterococci, Streptococcus pneumoniae, methicillin-resistant Staphylococcus aureus, vancomycin-intermediate or -resistant staphylococci, and Group A streptococci. The University of Iowa conducts the susceptibility testing, and the Hygienic Laboratory codes and serotypes the isolates. The data are available on the Internet so a health care practitioner can check on the prevalence of resistance in the area of the state where the patient resides.
Iowa is also planning a program that will detect infectious agents that could be released as a result of terrorism. Because such an attack is likely to result in the dispersal of infection in isolated settings throughout the country, the state public health facility is likely to be involved. In the various scenarios contemplated the need for adequate reporting and recognition by the attending physician are paramount for surveillance. For example, if a physician does not remember that anthrax is caused by a Bacillus species and does not notice that the laboratory reported it as a bacterial contaminant, he or she might not immediately attribute a death to anthrax. The cause of the disease might not be detected until later, when reports of a mysterious illness surface around the country. In some cases, the laboratory might presume that the isolate is a member of the patient’s normal flora and it is not until the isolate is referred to a central reference laboratory, possibly 2 weeks later, that the agent released by bioterrorists would be identified.
Identifying and reporting such agents are shared responsibilities of a national laboratory network of state facilities and local laboratories. In this regard, the Internet will serve as an invaluable tool for the sharing of information. In addition, what is needed is rapid communication, combined with algorithms for pathogen and disease identification, protocols for safety, a national laboratory training network, and the ability to detect multi-state outbreaks in real time. The Association of Public Health Laboratories recommends the formation of such a national network. State laboratories, because of their environmental chemical expertise, could augment the network by providing a means of detection of the agents of chemical terrorism. Preparation of state and private laboratories for bioterrorism events will enhance the ability to detect infectious diseases that emerge naturally.
Footnotes
- 1
The six specific diseases were tuberculosis, shigella toxin producing Escherichia coli infections, pertussis, cryptosporidiosis, hepatitis C, and penicillin-resistant streptococcal pneumonia. The diseases were chosen to represent various modes of transmission (foodborne, waterborne, and airborne) and different levels of antimicrobial resistance.
- 2
Data may be found in the GAO report issued after the workshop was held, titled, Emerging Infectious Diseases: Consensus on Needed Laboratory Capacity Could Strengthen Surveillance, (GAO/HEHS-99-26, February 5, 1999). A copy of this report may be obtained by calling (202) 512-6000, faxing a request to (202) 512-6061, or through the GAO website at http://www
.gao.gov (select GAO Reports and Testimony, select FY 1999 from the Annual Indexes, select the Authoring Division index, select Health, Education, and Human Services, choose HEHS-99-26). - 3
Infections refer to the entry and development of an infectious agent in the body of a person or animal. In an apparent, “manifest” infection, the infected person outwardly appears to be sick. In an unapparent infection, there is no outward sign that an infectious agent has entered the person at all. Infection should not be confused with disease.
- 4
Sepsis is the presence of pathogenic microorganisms or their toxins in blood or other tissues.
- 5
Meningitis is an inflammation of the membranes surronding the brain and spinal cord. People sometimes refer to it as spinal meningitis. Meningitis is usually caused by a viral or bacterial infection.
- 6
Encephalitis is an acute inflammatory disease of the brain due to direct viral invasion or to hypersensitivity initiated by a virus or other foreign protein.
- Surveillance - Public Health Systems and Emerging InfectionsSurveillance - Public Health Systems and Emerging Infections
Your browsing activity is empty.
Activity recording is turned off.
See more...