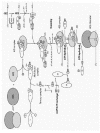
The initiation of protein synthesis consists in the recruitment of a ribosome·initiator tRNA complex to the initiation codon of a messenger RNA. In prokaryotes, this process involves the direct interaction of the ribosomal RNA with the mRNA. In contrast, eukaryotes have evolved a sophisticated mechanism that relies mostly on protein-RNA and protein-protein interactions. Eukaryotes have taken advantage of the evolution of novel mRNA structures, such as the 5' cap and the poly (A) tail, to develop new mechanisms for the recruitment of the ribosome to the mRNA. As a result, the eukaryotic translation initiation apparatus is now a complex machinery comprising at least eleven factors. This complexity provides a fertile ground for enhanced regulation, and many new mechanisms have been adopted by eukaryotes to control proteins synthesis. Indeed, many translation factors are phosphoproteins whose function can be regulated by extracellular signals. We will describe here the mechanism of translation initiation in eukaryotes, with a particular emphasis on translation factors and their function.
Introduction
With the structural confirmation that the ribosome is a ribozyme (see Chapter 15 by B.T. Wimberly), translation emerges as a process driven for the most part by RNA. As such, the initiation of protein synthesis appears to be devoted to the assembly of the catalytic rRNA and an initiator tRNA at the correct AUG codon of a template mRNA. These RNAs may now be widely acknowledged as the stars but they nevertheless cannot perform alone. Indeed the translation machinery, especially in eukaryotes, has evolved to require a host of protein factors.1 The focus of this review is on the essential parts played by the eukaryotic initiation factors (eIFs) in bringing together the initiator tRNA, ribosome and mRNA.
In eukaryotes, at least eleven different initiation factors are required to properly initiate translation.1 Collectively, they ensure that the methionyl-initiator tRNA (Met-tRNAiMet) is brought in the P site of the ribosome to the initiator AUG of an mRNA. Conceptually, this process can be divided in four steps:
- formation of the 43S pre-initiation complex, when the Met-tRNAiMet is delivered by eIF2 to the P site of the 40S ribosomal subunit;
- recruitment of the 43S complex to the 5' end of the mRNA by eIF3 and the eIF4 factors;
- scanning of the 5' untranslated region (UTR) and recognition of the AUG codon, and
- assembly of the 80S ribosome (Fig. 1).
Only the standard “scanning” mechanism utilized by most cytoplasmic mRNAs will be considered here. The reader is directed to recent reviews by Jackson2 and Hellen & Sarnow3 for a detailed discussion of alternative initiation pathways.
The sequence of several archaeal genomes revealed that translation in archaea is a hybrid between the bacterial and eukaryotic systems.4,5 Archaeal mRNAs are uncapped, polycistronic, lack long poly (A) tails, and their coding sequences are most often preceded by a Shine-Dalgarno element. In contrast to the bacterial mechanism for initiation codon selection, archaeal genomes contain homologues of many initiation factors that were thought to be exclusively eukaryotic.68 We will not discuss translation in archaea in more detail, except to point out that archaeal factors have been utilized in the last years to derive important structural and functional insights into the mechanisms of eukaryotic translation initiation (see below).
Formation of the 43S Preinitiation Complex
Initiator tRNA
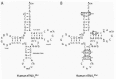
Translation is initiated by a special Met-tRNA in every organism.9 Because of its unique function, the initiator Met-tRNA (Met-tRNAiMet) presents many functional and structural features that distinguish it from the elongator Met-tRNA. In eukaryotes, the Met-tRNAiMet interacts specifically with an eIF2·GTP complex (see below), which delivers it exclusively to the ribosomal P site.10 This particular interaction can be explained by sequence divergence between the initiator and elongator form of Met-tRNA (Fig. 2). Some sequence specificities promote the activity of the Met-tRNAiMet in initiation (Fig. 2). These include the A1:U72 base pair which is critical for initiator function and cell viability in S. cerevisiae11 and, when mutated to the G1:C72 present in elongator Met-tRNA, weakens the affinity of the human Met-tRNAiMet for eIF2 by a factor of 10.12 The critical function of the A1:U72 base pair in initiation is underscored by its almost universal conservation amongst cytoplasmic eukaryotic initiator tRNAs, and its exclusion from all eukaryotic elongator tRNAs.13 In addition, the Met-tRNAiMet loop IV contains A60 in place of pyrimidine 60, and A54 instead of the T54 present in the TγC sequence found in elongator tRNAs (Fig. 2). The presence of an A at positions 54 and 60 is not absolutely required, as only the A54U substitution is lethal in yeast.11 In comparison, the double substitution A54, A60 to T54, U60 in the human tRNA has minimal effect on an in vitro initiation assay.14 A yeast Met-tRNAi Met with both G1:C72 and T54 mutations rescues an elongator tRNAMet-depleted strain, suggesting a requirement of these residues for initiator functions and discrimination against elongator tRNAMet.15 Finally, the Met-tRNAiMet contains three consecutive G:C base pairs in the anticodon stem (Fig. 2). Substituting these nucleotides with those found in the elongator tRNA reduces the initiator activity in vitro.14 Furthermore, introduction of G29:C41 and G31:C39 in a yeast elongator tRNA with the A1:U72 and A54 mutations increases its activity in initiation.15
Other sequence specificities are important for the discrimination of initiator and elongator tRNAMet (Fig. 2). Position 64 in plants and fungi has a unique 2'-O-phosphoribosyl group, which disrupts the structure of the TψC stem.9,16 This modification prevents the elongator function of Met-tRNAiMet,17,18 probably by inhibiting its interaction with the eEF1A·GTP complex.19,20 In vertebrates, it has been proposed that the TγC stem perturbation is due to the nature of the base pairs at the base of the stem.21 Indeed, mutation in base pairs A50:U64 and U51:A63 yield an initiator tRNA that participates in elongation and, when combined with the A1:U72 -> G1:C72 mutation, results in almost wild-type elongator function.21
eIF2
The initiator tRNA is delivered directly to the P site of the 40S ribosomal subunit as part of a ternary complex that also contains the heterotrimeric G protein eIF2 and GTP (Fig. 1). Subsequent pairing between the AUG initiation codon and the Met-tRNAiMet anticodon elicits hydrolysis of the GTP by eIF2, which requires the GTPase activating protein (GAP) eIF5 (Fig. 1; see below). The eIF2·GDP complex does not bind to the Met-tRNAiMet, and recycling of the GTP necessitates the guanine nucleotide exchange factor (GEF) eIF2B (Fig. 1; see below). An eIF2 homologue is present in archaea but not in bacteria, where IF2 delivers the fMet-tRNAfMet to the 30S subunit.7
Essential genes encode the α, β and γ subunits of eIF2 in S. cerevisiae. Mutations in all three subunits affect the accuracy of AUG initiation codon selection.22 Biochemical and genetic analyses indicate that eIF2γ likely binds to GTP and Met-tRNAiMet.10,22 Interaction between eIF2γ and the initiator tRNA is reduced by the Y142H and N135K mutations in yeast.23,24 The N135K mutant also increases GTP hydrolysis,24 whereas the K250R mutation increases the off-rate for GDP and GTP.23 These observations have recently been corroborated by the crystal structure of the archaeal eIF2γ (a-eIF2γ) from Pyrococcus abyssi.25 Sequence similarity7 suggests that a-eIF2γ is a close structural homologue of the elongation factors eEF1A and the bacterial EF1A (see Chapter 20 by P. Nissen et al and ref. 25). This is most apparent in the nucleotide-free structure, which shows that the three domains of a-eIF2γ are very similar to their equivalents in eEF1A and EF1A, although their relative orientation is somewhat different.25 Accordingly, the GDP binding pocket of a-eIF2γ is identical to that of EF1A. Superimposition of domains 2 and 3 from the EF1A·GTP·Phe-tRNAPhe structure on the corresponding domains in a-eIF2γ indicates that the Phe-tRNAPhe can readily be accommodated by the structure, except for the Phe side chain.25 This in turn suggests a binding pocket for the Met side chain of Met-tRNAiMet, which might explain why the methionyl group enhances initiation.26 The structure also shows that a-eIF2γ lacks features allowing EF1A to bind aminoacyl-tRNA nonspecifically, helping to explain the exclusion of elongator tRNA from eIF2.25 Unfortunately, no difference could be observed when comparing the GDP-bound structure of a-eIF2γ to the GMP-PNP bound form, which might indicate that eIF2β has a role in tRNAi Met binding to eIF2. Therefore, the structural explanation of how the hydrolysis of GTP causes the release of the Met-tRNAiMet will probably require a co-crystal of a-eIF2γ with a-eIF2β, a-eIF2α or both, along with the initiator tRNA. An interesting aspect of the a-eIF2γ structure is the occurrence of a conserved four cysteine cluster that was found to coordinate a zinc atom.25 The specific role of this motif is not known, but its significance has been highlighted in yeast where mutating the second cysteine (C155) to a serine causes a slow growth phenotype.27
While eIF2γ forms the structural core of eIF2, essential functions are also performed by eIF2β.10 Genetic screening in yeast identified eIF2β mutations in an essential Cys2-Cys2 zinc-finger motif found in the carboxy-terminal domain of eIF2β.22 Two of these mutants (S264Y and L254P) were biochemically characterized and confer an intrinsic GTPase activity to eIF2, implicating eIF2β in promoting eIF2γ GTPase activity.24 Independent of GTP hydrolysis, the S264Y mutant also increases initiator tRNA dissociation from eIF2.24 In addition, the zinc-finger motif is part of a larger mRNA binding region, and could therefore be important for facilitating codon-anticodon interaction in eIF2.28 mRNA binding properties were also ascribed to the three lysine repeats found at the amino-terminus of eIF2β.28 Moreover, these repeats mediate the mutually exclusive interaction of eIF5 and eIF2Bϵ with eIF2β.29,30 In agreement with the absence of eIF2B and eIF5 in archaea, a-eIF2β does not possess the region containing the lysine repeats. The solution structure of a-eIF2β from Methanococcus jannaschii was recently solved.31 a-eIF2β consists of two independent structural domains connected by a conserved helical linker.31 The N-terminal domain of a-eIF2β corresponds to the central domain of yeast eIF2β, which is responsible for the interaction with eIF2γ.32 The structure of the C-terminal domain reveals a critical role for the zinc atom in maintaining the structural integrity of the domain.31 Yeast eIF2β mutations implicated in initiation codon selection22 affect residues that are conserved in a-eIF2β. Four of these residues (Lys122, Arg125, Val 126 and Ile140) are located in close proximity in the structure of the C-terminal domain,31 and might serve as a binding site for other translational components implicated in initiation codon recognition.31
Phosphorylation of the conserved serine residue (Ser51) in eIF2α converts eIF2·GDP from a substrate to a competitive inhibitor of eIF2B.10 Whereas eIF2α is required for yeast survival under normal conditions, it is not essential as long as the β and γ subunits as well as the initiator tRNA are overexpressed.33 Removal of the α subunit of eIF2 causes an 18-fold increase in the Km of eIF2B catalyzed nucleotide exchange, indicating that eIF2α is required for interactions between eIF2 and eIF2B that promote wild-type rates of nucleotide exchange.34 Consistent with this, phosphorylation of eIF2α on Ser51 increases its affinity for the regulatory subcomplex of eIF2B (α, β, δ), in a way that hampers the function of the eIF2B catalytic subcomplex (γ, ϵ).35 Taken together, these data clearly indicate that the a subunit of eIF2 has evolved to become first and foremost a regulator of translation initiation. The X-ray structure of the N-terminal two-thirds of human eIF2α has been solved.36 This structure contains two domains: an N-terminal oligonucleotide binding (OB) domain similar to that in IF137 and eIF1A,38 and a C-terminal helical domain.36 The Ser51 residue is located in an unstructured loop connecting β3 and β4 of the OB domain. The putative RNA binding site of the OB domain lacks the cluster of positive charges that is observed in other members of the OB family, which is consistent with eIF2α not interacting with RNA.36 Protein-protein interactions involving eIF2α could be mediated by a highly conserved groove that is observed between the OB and helical domains.36
eIF2B
eIF2B is a heteropentameric GEF that is required to recycle the inactive eIF2·GDP complex into eIF2·GTP (Fig. 1). eIF2B is present in cells in lower amounts than eIF2.39 Phosphorylation of eIF2 on Ser51 of its α subunit sequesters eIF2B, thus repressing translation initiation.10 Genetic and biochemical methods indicate that eIF2B is divided into a catalytic subcomplex comprising eIF2Bγ and eIF2Bϵ, and a regulatory subcomplex consisting of eIF2Bα, eIF2Bβ and eIF2Bδ.40 The catalytic subcomplex can perform the guanylate exchange reaction even when eIF2α is phosphorylated, unless the regulatory subcomplex is present to modulate its activity.40
eIF2Bϵ and eIF2γ display sequence homology throughout their length, and both subunits are essential in yeast.10 eIF2Bγ does not exhibit catalytic activity, but it enhances the weak intrinsic guanylate exchange activity of eIF2Bϵ.40,41 Indeed, the catalytic subcomplex exhibits a higher activity than the holoenzyme, probably owing to the absence of regulation from the other subunits.40 The catalytic site is located in the C-terminal region of eIF2Bϵ, and is independent from the bipartite motif that is required for binding to eIF2β.30,42 The N-terminus of eIF2Bϵ contains elements that are necessary for the assembly of eIF2Bϵ and eIF2Bγ into the eIF2B complex.42
eIF2Bα, eIF2Bβ and eIF2Bδ also display sequence homology to each other. These three subunits are required for the regulatory subcomplex to bind to eIF2α and differentiate between its phosphorylated and nonphosphorylated forms.40 eIF2Bβ and eIF2Bδ are essential in yeast, but eIF2Bα is dispensable.10 However, the absence of eIF2Bα renders eIF2B insensitive to the phosphorylation status of eIF2α, both with yeast and rat factors.40,43 In summary, eIF2B appears to have evolved to function in two roles: a GEF to recycle eIF2·GDP, and a regulator to respond to eIF2α phosphorylation.
Interestingly, mutations in the five subunits of human eIF2B are associated with leukoencephalopathy with vanishing white matter (VWM), a rare brain disease usually affecting children.44,45
This is the first documented disease to be associated with mutations in a translation factor. Many missense mutations were observed in the different eIF2B subunits, but nonsense mutations were always found to be heterozygous.44,45 These data suggest that mutations completely inactivating eIF2B are lethal when homozygous, in agreement with the essential role of eIF2B.10 The central nervous system specificity of the disease is surprising. Translational regulation by eIF2B involves the phosphorylation of eIF2 on the a subunit in response to various stress signals (see below). Thus, it is possible that the brain is more sensitive to an altered stress response brought about by mutated eIF2B. Indeed, neurological deterioration is observed during or after episodes of fever in VWM patients.44,45
eIF3
The largest and most complex initiation factor, eIF3, is composed of up to 11 nonidentical subunits in mammals, 6 of which are conserved in yeast (Table 1).1,10 In agreement with its elaborate constitution, eIF3 has multiple functions in translation initiation:
- it is thought to promote the dissociation of 40S and 60S ribosomal subunits,
- it participates in the recruitment of the ternary complex to the 40S subunit, and
Table 1
eIF3 subunit composition.
eIF3 and eIF1A are thought to promote ribosome dissociation, but the mechanism is still poorly characterized.1 Recent work suggests that eIF3 prevents 60S subunits from disrupting the 43S preinitiation complex.46 This is consistent with earlier observations of eIF3 being associated with native 40S subunits,47 which implies that the ternary complex binds to a preformed 40S·eIF3 complex. Indeed, a ternary complex bound to 40S subunits in the absence of eIF3 was readily dissociated by 60S subunits.46 Thus, eIF3 appears to be required for the stabilization of ternary complex binding to the small ribosomal subunit.
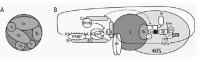
The study of eIF3 in yeast has recently clarified its role in the recruitment of the ternary complex to 40S subunits. eIF3 from S. cerevisiae is composed of a core of five essential subunits that are conserved in mammals (Table 1): eIF3a, eIF3b, eIF3c, eIF3g and eIF3i.10,48,49 The core complex co-purifies with eIF5,48 an interaction that has also been observed in mammalian cells.50 This interaction is mediated by the amino-terminus of eIF3c,30,51 and is important for bridging eIF3 to eIF2, since both can concurrently bind to the C-terminal bipartite motif of eIF5 (Fig. 3).51 eIF1 can also interact with eIF3c,49,52 and a large multifactor complex consisting of eIF1/eIF2/eIF3/eIF5/ Met-tRNAiMet is observed in vivo in the absence of 40S subunits (Fig. 1).51 Since eIF1, eIF5 and the three eIF2 subunits promote stringent AUG selection,10,22,24 eIF3 could potentially have a structural function in AUG recognition. Thus, the multiple protein-protein interactions mediated by eIF3 appear to be critical in the recruitment or stable binding of the ternary complex to 40S subunits (Fig. 3).
A yeast homologue of the mammalian eIF3j is not associated with the purified eIF3 core complex. 48 However, it appears that at least a fraction of eIF3j is incorporated into eIF3.53 Like its mammalian counterpart,54 yeast eIF3j interacts with eIF3a.53 Interestingly, the amino acid sequence of eIF3j is similar to the middle region of eIF3a, and forms a motif that is sufficient for binding to the RNA recognition motif (RRM) of eIF3b. Deletion of eIF3j is not lethal but causes a destabilization of eIF3 and its associated factors, suggesting that it is important in stabilizing the interaction of eIF3a with eIF3b.53 The latter interaction is critical in yeast, since deletion of the eIF3b RRM releases eIF3a and induces the breakdown of the multifactor complex, with the remaining eIF3b-eIF3g-eIF3i being unable to bind 40S subunits.53 In support of these observations, a purified subcomplex of eIF3 containing eIF3a, eIF3b and eIF3c is found to integrate eIF3j, eIF1 and eIF5, and can rescue translation in vitro almost as efficiently as purified eIF3 core complex.55
Finally, eIF3 is required for the recruitment of mRNA to the 43S preinitiation complex. In mammals, this is thought to be due to the interaction of eIF3 with eIF4F, which acts as a bridge to the mRNA through the interaction of eIF4E with the cap structure.56,57 The subunit of eIF3 responsible for this interaction has not been identified yet, and there is no report of a similar interaction in yeast. However, it was recently reported that yeast eIF5 can interact with the carboxy-terminal half of eIF4G, and could act as a link between eIF3 and the mRNA (Fig. 3).58 Another factor that could be involved in recruiting the 43S preinitiation complex to the mRNA is eIF4B (see below), which interacts with eIF3a in mammals,59 and with eIF3g in S. cerevisiae.60
Recruitment of the 43S Complex to the mRNA
Cap Structure
Eubacterial and archaeal mRNAs possess a Shine-Dalgarno sequence that facilitates ribosome recruitment by interacting directly with the complementary sequence at the 3' end of the 16S ribosomal RNA. Despite having a very similar 3' end, the eukaryotic 18S rRNA lacks such an element.2 To identify the 5' end of mRNAs, eukaryotic cells have instead opted for a specific structure called a cap. This m7GpppX sequence (where X is any nucleotide) is present at the 5' end of all nuclear transcribed mRNAs from eukaryotes,61 and has possibly evolved to stabilize mRNAs. The cap facilitates translation initiation, but also participate in other processes such as mRNA splicing and nucleocytoplasmic transport.62 The recruitment of the 43S complex to the mRNA is mediated by the eIF4 group of factors. The function of these factors has been thoroughly reviewed elsewhere,63 and thus only a summary of our current understanding will be presented here.
eIF4E
eIF4E interacts with the cap structure and, together with eIF4G and eIF4A, forms the eIF4F complex (Fig. 1). eIF4E is an essential factor that is conserved across all eukaryotic kingdoms. The structures of yeast and mouse eIF4E bound to m7GDP were solved,64,65 and demonstrate the conservation of the cap-binding mode. Indeed, all the amino acids involved in cap recognition are conserved from yeast to humans, which strongly suggests a common ancestral origin. In light of the fact that an eIF4E homologue is absent from archaea, it will be very interesting to address the question of the origin of this factor.66 Why did the eukaryotes select to use the cap structure to initiate translation? What selective advantage is achieved by adopting such a radically different ribosome recruitment mode?
In mouse and human, eIF4E is phosphorylated on one major site, serine 209.67 The function of this phosphorylation in not completely understood. Phosphorylation of eIF4E is correlated with increased translation rates, but it is still disputed whether phosphorylation increases the cap binding affinity of eIF4E.6870 The structure suggests that phosphorylated Ser209 could form a salt bridge with Lys159, which would act as a clamp above the proposed trajectory of the mRNA path.64 However, a more recent structural analysis of the full-length eIF4E complexed with m7GpppA indicates that the distance between Ser209 and Lys159 would be too long to form a salt bridge.68 In fact, the C-terminal loop region of eIF4E, which encompasses Ser209, interacts with the second nucleotide of the mRNA. Hence, phosphorylation of Ser209 could still have an effect on the affinity of eIF4E for capped mRNA.68
Notwithstanding its structural basis, the biological significance of eIF4E phosphorylation is not immediately clear. Studies on eIF4E phosphorylation mutants have demonstrated that they can restore translation as well as the wild-type protein in an in vitro assay.71 These eIF4E variants can also rescue the lethal phenotype of eIF4E deletion in yeast.71 In contrast, transgenic Drosophila melanogaster expressing an eIF4EI phosphorylation mutant (S251A, analogous to S209A) displays reduced viability (65% survival) in an eIF4EI mutant background, with the surviving flies having growth retardation defects.72 These defects were not observed when a S251D mutant was used to mimic a constitutively phosphorylated residue.
eIF4E is phosphorylated in vivo by the kinase Mnk1.73,74 In order to phosphorylate eIF4E, Mnk1 needs to be recruited to the C-terminal part of eIF4G (Fig. 3).75 It appears highly unlikely that such a sophisticated mechanism would have evolved if it did not have a purpose. Therefore we conclude that eIF4E phosphorylation must play an important regulatory role in vivo, at least in higher eukaryotes. This will probably be more fully appreciated by performing in vivo studies of a mouse knock-in bearing an eIF4E Ser209Ala mutation.
eIF4G
Two related proteins, eIF4GI and eIF4GII, function as modular adaptor proteins that mediate the series of protein-protein interactions culminating in the recruitment of the 43S complex to the mRNA 5' end (Fig. 3).63,76,77 eIF4G has evolved around a phylogenetically conserved central core (Fig. 4).66 This core is contained within the middle fragment of the protein and possesses two interaction domains: the first interacts with the eIF4A RNA helicase,78,79 and the second is an RNA binding domain.80,81 In addition, this fragment of mammalian eIF4G also contains an eIF3 interaction domain.56,78 The X-ray structure of the middle portion of human eIF4GII has been solved, and exhibits a crescent-shaped domain consisting of ten alpha helices organized into five HEAT repeats. 82 Proteins containing HEAT repeats are involved in the assembly of large multiprotein complexes, which is probably facilitated by the large and accessible surface area of the domain.83
Extension of the central core of eIF4G to include the eIF4E-binding site generates an eIF4G domain that is necessary and sufficient for cap-dependent translation initiation.84 The 4E-binding site is apparently only required for targeting eIF4G to the mRNA. Indeed cap-independent translation of an mRNA containing an iron response element (IRE) can be directed by the central core of eIF4G, if this domain is fused to the IRE-binding protein IRP-1.85,86
The amino-terminal portion of eIF4G also contains an interaction domain for the poly(A) binding protein (PABP).87,88 PABP is a cytoplasmic protein that binds to the poly(A) tail present at the 3'-end of messenger RNAs. Its interaction with eIF4G provides a physical link between the cap and the poly(A) tail (Fig. 3). It also explains the observation that the poly(A) tail can operate as an enhancer of translation initiation.89,90 The significance of the cap-poly(A) tail interaction is discussed in more detail in Chapter 12.
The C-terminus of eIF4G has been considerably extended during the expansion of eukaryotes.66 A second eIF4A interaction domain is present in human eIF4G.78 This domain is absent from yeast eIF4G, but can be found in Arabidopsis and Drosophila eIF4G homologues. The related protein p97/NAT1/DAP-5 has a similar domain,66 which on the other hand does not bind to eIF4A.78 In eIF4G, the C-terminal eIF4A binding domain is not essential for translation, but instead plays a role in modulating the function of the core domain.84 There is no agreement as to whether eIF4G interacts with one or two eIF4A molecules.84,91,92 The extreme C-terminus of eIF4G from metazoans displays another conserved motif which, in mammals, is necessary for the interaction with the eIF4E kinase Mnk1.75 It is intriguing that this motif is conserved in the C-termini of eIF5 and eIF2Bϵ,93 and participates in their interaction with eIF2β.30 This may represent a conserved module important for protein-protein interactions regulating the assembly of the translational apparatus.66
eIF4A
eIF4A is a member of the large family of RNA helicases termed DEAD-box, after one of their 8 conserved motifs.94 In yeast, eIF4A is encoded by two genes whose simultaneous inactivation is lethal.95 There are three eIF4A isoforms in mammals, but only eIF4AI and eIF4AII appear to be involved in translation initiation.96 Interestingly, mammalian eIF4A can interact with yeast eIF4G1 in vitro,79 even if it cannot complement yeast mutants defective in eIF4A.97 In fact mammalian eIF4A appears to act as a translational inhibitor in yeast,79 much like eIF4AIII inhibits translation in mammals.96 eIF4A is the only eIF4 factor to have a homologue in archaea.6
eIF4A is an RNA dependent ATPase and can unwind RNA duplexes by itself or, much more efficiently, as part of the eIF4F complex.63 The intrinsic helicase activity of eIF4A is relatively weak, but it is strongly stimulated by eIF4B, although the two proteins do not appear to interact directly.98 Recent studies suggest that eIF4A is a nonprocessive helicase that is limited to a single round of unwinding.99,100 The presence of eIF4B (or eIF4H) may limit RNA re-association and allow the unwinding of longer duplexes.99,100 It is noteworthy that eIF4A functions primarily as part of the eIF4F complex, but that it apparently needs to be exchanged with free eIF4A to effect unwinding.101 It is possible that this recycling process is what conveys processivity to the complex. An unanswered question is how does eIF4A facilitate ribosome bindingγ eIF4F is generally believed to target eIF4A to the mRNA, which then unwinds secondary structures to allow ribosome recruitment. The role of eIF4A could be to utilize the energy from ATP hydrolysis to rearrange RNA structures, or maybe RNA-protein complexes, thus allowing 40S subunit binding to the mRNA.102,103
eIF4B
The function of eIF4B in translation initiation is not completely clear. In vitro, eIF4B enhances the helicase activity of eIF4A and eIF4F,98 and promotes 48S complex assembly.84,104 However, eIF4B is not required in ribosome binding assays,84,104 and yeast strains with a disrupted eIF4B gene display translational defects and a slow-growth phenotype, but are viable.105,106
eIF4B homodimerizes and interacts with RNA through a N-terminal RRM and a C-terminal arginine-rich motif (ARM).59,107,108 Since eIF4B also demonstrates an RNA-annealing activity, it was proposed that it could play a role in mediating interactions between the rRNA and mRNA.109 Consistent with this hypothesis, eIF4B can interact with the 18S rRNA in vitro, and can simultaneously bind to two RNA molecules.110
eIF4H
eIF4H is a recently identified factor that shares functional and sequence homology with eIF4B, mostly with the RRM domain.111 It weakly binds RNA and stimulates protein synthesis in vitro, as well as the ATPase and helicase activity of eIF4A and eIF4F.99,100,111,112 A recent report shows that the virion host shutoff protein (Vhs) of herpes simplex virus interacts with eIF4H.113 This protein accelerates mRNA degradation, which modulates the levels of viral and cellular gene expression. It appears that Vhs is targeted to the mRNA 5' end through its interaction with eIF4H.113
Scanning and Localization of the Initiator AUG
mRNA 5' UTR
After binding to the 5' end of the mRNA, the preinitiation complex must locate the AUG codon. It is believed that the ribosome and associated factors migrate through the 5' UTR until the Met-tRNAiMet can form a productive base pairing with an initiation codon. This has been defined as the scanning model,114,115 and is consistent with the experimental data. For example, the insertion of an upstream AUG usually creates a new translation start site. In addition, insertion of a stable secondary structure in the 5' UTR dramatically reduces translational efficiency, probably by hampering ribosome movement.116 The minimal set of factors required for 48S complex assembly at an initiation codon has been better defined in recent years (see below).117,118 Nevertheless, scanning has never been biochemically assayed and little is known about its mechanism.
eIF1
Sequence homologues of eIF1 have been identified in archaea and bacteria, indicating an ancient origin for this factor.8 In yeast, eIF1 is essential for viability,119 and genetic screenings have identified a number of mutations affecting the fidelity of initiation codon selection.22 These mutations affect residues that are conserved between eukaryotes, archaea and bacteria,8 and the NMR structure of eIF1 suggest that they represent an interaction surface for other molecules.52 A fraction of eIF1 from S. cerevisiae copurifies with eIF3, through an interaction with eIF3c.48,49,120 This might explain how eIF5 mutants can suppress the effect of eIF1 mutations in yeast,22 because eIF3c can interact simultaneously with eIF1 and eIF5 (Fig. 3).51 Thus, eIF1 is in close proximity to eIF5 and could affect its GTPase activating function. Toe-printing analysis of pre-initiation complexes bound to mRNA have provided evidence that eIF1 is required for the formation of 48S complexes.117 In this assay, which also requires eIF2, eIF3, eIF4A, eIF4B, eIF4F and Met-tRNAiMet, eIF1 can weakly promote the correct positioning of 43S complex at the initiation codon. This activity is strongly enhanced by eIF1A (see below).117 The molecular mechanism remains however unclear, and thus the exact function of eIF1 could be to participate in scanning, destabilize incorrectly positioned complexes, or stabilize correctly positioned ones.
eIF1A
eIF1A is the eukaryotic homologue of the bacterial IF1 and the archaeal a-eIF1A.8 The 3D structures of eIF1A shows that it contains an OB domain similar to that of IF1.37,38 eIF1A is essential in S. cerevisiae and appears to have multiple roles in translation initiation.1 Pestova et al reported that eIF1A acts in synergy with eIF1 to promote the assembly of 48S complex at the initiation codon.117 However, eIF1A without eIF1 can only promote the formation of an aberrant complex located close to the cap. This cap-proximal complex is not a translational intermediate, and it is unable to reach the AUG codon.117 eIF1A also promotes the binding of the ternary complex to isolated 40S subunits.46,121 In addition, eIF1A interacts with eIF5B.122 Along with structural studies of a 30S·IF1 complex,123 this suggests that eIF1A occupies the A site of the small ribosomal subunit and, in conjunction with eIF5B, directs the initiator tRNA to the P site.124
60S Ribosomal Subunit Joining
Once the preinitiation complex has reached an initiator codon, base pairing between the AUG and the Met-tRNAiMet anticodon elicits a series of events that culminate in the joining of the 60S ribosomal subunit to form an active ribosome that is competent for elongation. This involves the release of the initiation factors bound to the 40S subunits, which requires GTP hydrolysis. Recruitment of the 60S subunit is not spontaneous after the factors release, and it necessitates additional initiation factors.
eIF5
Once the 48S complex is correctly assembled at the initiation codon, the bound initiation factors must be displaced to allow the joining of the 60S subunit. This step requires the hydrolysis of the GTP molecule bound to eIF2 (Fig. 1), since its substitution with the nonhydrolyzable GMP-PNP arrests initiation at the 48S stage.125 Activation of the GTPase function of eIF2 requires its association with eIF5 and the 40S subunit.10 eIF5 is essential in yeast, and an activated eIF5 mutant (G31R) can initiate translation at non-AUG codons.22 This mutant demonstrates a two-fold stimulation of GTP hydrolysis. Combined with the studies on eIF2 mutants showing intrinsic GTPase activity, it appears that eIF2 and eIF5 act in concert to maintain the accuracy of initiation codon recognition in eukaryotes. Intriguingly, eIF5 has not been found to interact with the GTP-bound eIF2γ, but rather with eIF2β, at the same site which was demonstrated to interact with eIF2Bϵ.29,30 This argues that GTP hydrolysis must be triggered by a conformational change in eIF2, when the initiator tRNA is correctly paired with the AUG codon. The amino-terminus of eIF5 is similar to the carboxy-terminus of eIF2β, including the Cys2-Cys2 zinc-finger motif, and is expected to adopt a similar fold.31
Recent observations suggest that eIF5 acts as a bridge between eIF3 and eIF2, and would therefore participate in the recruitment of the ternary complex to the 40S subunit (see above).51 It has also been demonstrated that yeast eIF5 can interact simultaneously with eIF3 and eIF4G, and could thus take part in 43S complex recruitment to the mRNA.58 These interactions are all mediated by the C-terminal domain of eIF5, while the GTPase activating region apparently resides in the N-terminus.58 Interestingly, the amino-terminus of eIF5 shares sequence homology with the carboxy-terminal fragment of eIF2β.29,31
eIF5B
eIF5B was first identified as a translation factor in yeast, and termed yeast IF2 (yIF2) for its homology to the prokaryotic IF2.126 Human,127,128 Drosophila,129 and archaeal7 homologues were also identified, making IF2/eIF5B a universally conserved translation factor. Disruption of the eIF5B gene (FUN12) in yeast causes a severe slow-growth phenotype, associated with a defect in translation initiation.126 It was later demonstrated that eIF5B has a function analogous to prokaryotic IF2 in mediating the joining of the 60S ribosomal subunit.130 The requirement for both eIF2 and eIF5B strongly suggest that translation initiation in eukaryotes requires the hydrolysis of two GTP molecules (Fig. 1).125 This is consistent with the kinetic analysis of translation initiation in vitro,131 and offers additional possibilities for regulation.
The structure of eIF5B has revealed an unusual architecture.132 It consists of three N-terminal domains (I, II, III) connected by a long a helix to domain IV. Domain I is a G domain, domains II and IV are β-barrels and domain III has a novel a/β/a sandwich fold.132 The G domain and the β-barrel domain II display a similar structure and arrangement to the homologous domains in EF1A, eEF1A and a-eIF2γ.25,132 This suggests that they form a core structure present in all GTPases involved in translation.124 GTP-bound eIF5B facilitates 60S subunit joining, but GTP hydrolysis occurs after 80S formation and is required for the release of eIF5B.130 The comparison of eIF5B·GTP and eIF5B·GDP demonstrates that, like other GTPases, GTP hydrolysis induces a modest conformational switch in the G domain. This small modification is however amplified via a coordinated rearrangement of domains II-IV, resulting in the movement of domain IV.132 Domain IV is essential for the in vivo function of eIF5B and interacts with eIF1A, suggesting that the release of eIF1A and eIF5B from the ribosome could be coupled.122
Regulation of Translation Initiation
As with any multi-step process, eukaryotic translation can be regulated at various levels. However, it is generally more efficient to regulate complex pathways at their initiation step, and this is where translational control most often occurs.133 Two factors are known to play a critical role in the regulation of translation initiation: eIF2 and eIF4E. The activity of eIF2 is controlled by the rate of GTP recycling, and thus by the availability of eIF2B.10 The function of eIF4E is regulated by its incorporation into eIF4F, which is inhibited by the eIF4E-Binding Proteins (4E-BPs).63 Both of these mechanisms are subjected to regulatory phosphorylation and can thus provide rapid and reversible means to maintain cellular homeostasis in response to environmental signals. We will give a brief outline of how the activity of eIF2 and eIF4E can be regulated. The reader is directed to a recent review by Dever134 for a more detailed discussion of how gene-specific regulation can be achieved by modulating the activity of general translation factors. An in depth review of the multiple translational control mechanisms can be found in reference 135.
eIF2α Kinases
As mentioned above, the phosphorylation of eIF2α on Ser51 results in the conversion of eIF2 from a substrate to a competitive inhibitor of eIF2B. The critical importance and versatility of this regulatory event is emphasized by the successive appearance of the four known eIF2• kinases through the eukaryotic lineages: mammals acquired the double-stranded RNA-activated protein kinase (PKR); vertebrates, the hemin-regulated inhibitor kinase (HRI); metazoans, the PKR-like ER kinase (PERK); while all eukaryotes possess the kinase GCN2.10,136138 These four kinases are activated by environmental stress.139 PKR is part of the anti-viral response, since its expression is induced by interferon and its kinase function is activated by double-stranded RNA.138 HRI regulates protein synthesis in erythroid cells in response to heme deprivation, heat shock and oxidative and osmotic stress.137,140 PERK is activated as part of the unfolded protein response, which is caused by stress to the endoplasmic reticulum.136 Finally the activity of GCN2 can be stimulated by a variety of conditions, the best characterized being amino acid starvation.10,141
eIF4E-Binding Proteins
The complex m7GpppX·eIF4E·eIF4G directs the 43S preinitiation complex to the mRNA 5' end. The 4E-BPs specifically inhibit cap-dependent translation initiation by preventing the interaction of eIF4E with eIF4G.63 Three 4E-BPs were identified and found to have similar functions,142,143 but 4E-BP1 has been more thoroughly characterized. The binding of 4E-BP1 to eIF4E is reversible, and is regulated by the phosphorylation state of 4E-BP1. Upon stimulation of cells with serum, growth factors or hormones, 4E-BP1 becomes phosphorylated on specific serine/threonine residues and dissociates from eIF4E to relieve translational inhibition.63,144 Conversely, cellular stresses such as nutrient deprivation or viral infections will cause a decrease in 4E-BP1 phosphorylation and increase its affinity for eIF4E.63,67 The phosphorylation events leading to the dissociation of 4E-BP1 from eIF4E are mediated by the FKBP12-rapamycin associated protein/mammalian target of rapamycin (FRAP/mTOR), and probably other kinase(s) that have yet to be identified.144146 The activity of FRAP/mTOR is modulated by the availability of nutrients such as amino acids, and by the intracellular concentration of ATP.146148 Given that FRAP/mTOR signals not only to 4E-BP1 but to other translational targets (eIF4B, eIF4GI, eEF2 and S6K1), it emerges as a molecular sensor capable of integrating diverse inputs to modulate protein synthesis in response to a cell's nutritional and energetic status.146,148
Conclusion
The structural and functional analysis of eukaryotic translation initiation factors reveals that they have most likely evolved to take advantage of the many RNA structural elements that are specific to eukaryotes. Archaeal translation illustrates the evolution of the protein synthesis machinery from eubacteria to eukaryotes, exhibiting characteristics from both systems. For example, archaeal mRNAs have a Shine-Dalgarno sequence but utilize a factor homologous to eIF2 for Met-tRNAiMet delivery to the ribosome. Many of the eukaryotic translation initiation factors do not have counterparts in prokaryotes, and must have evolved together with the unique features of eukaryotic mRNA (cap structure, poly(A) tail). Archaea lack a 5' cap or long poly(A) tail and, accordingly, are devoid of factors related to eIF3, eIF4G or eIF4E. It can therefore be envisaged that eIF4G has evolved through the accretion of various interaction domains around a conserved central core. An eIF4E-binding site could thus have been added after the 5' cap structure has evolved, and a PABP interaction domain after the poly(A) tail was developed. Thus, RNA structural elements appear to have played a critical role in directing the evolution of the eukaryotic translational machinery. The increase in complexity that has resulted served to develop new regulatory mechanisms to control gene expression, such as the sequestration of eIF4E by the 4E-BPs or the phosphorylation of eIF2α. Once evolved, regulatory mechanisms could be subjected to further refinement. For instance, mammals have evolved four different eIF2α kinases. As a consequence of the increased regulatory capabilities, translation initiation has become a very dynamic process that can be delicately and rapidly modulated in response to environmental changes. Thus, the ability to better control translation is likely to have provided eukaryotes with increased adaptive flexibility.
Acknowledgements
We would like to thank Thomas Dever, Emmanuel Petroulakis, Mathieu Miron and Park Cho-Park for critical reading of the manuscript. F.P. is supported by a Doctoral Award from the Canadian Institutes of Health Research (CIHR). N.S. is a CIHR Distinguished Investigator and a Howard Hughes Medical Institute International Scholar.
References
- 1.
- Hershey JWB, Merrick WC. The pathway and mechanisms of initiation of protein synthesisIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,200033–88.
- 2.
- Jackson RJ. A comparative view of initiation site selection mechanismsIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000127–183.
- 3.
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. [PubMed: 11445534]
- 4.
- Bult CJ, White O, Olsen GJ. et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. [PubMed: 8688087]
- 5.
- Klenk HP, Clayton RA, Tomb JF. et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. [PubMed: 9389475]
- 6.
- Dennis PP. Ancient ciphers: translation in Archaea. Cell. 1997;89:1007–1010. [PubMed: 9215623]
- 7.
- Kyrpides NC, Woese CR. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B alpha-beta-delta subunit families. Proc Natl Acad Sci USA. 1998;95:3726–3730. [PMC free article: PMC19904] [PubMed: 9520434]
- 8.
- Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–228. [PMC free article: PMC18182] [PubMed: 9419357]
- 9.
- RajBhandary UL, Chow CM. Initiator tRNAs and initiation of protein synthesisIn: Söll D, RajBhandary UL, eds.tRNA: Structure, Biosynthesis, and FunctionWashington, DC: American Society for Microbiology;1995511–528.
- 10.
- Hinnebusch AG. Mechanism and regulation of initiator Methionyl-tRNA binding to ribosomesIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000185–243.
- 11.
- von Pawel-Rammingen U, Astrom S, Bystrom AS. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1432–1442. [PMC free article: PMC369584] [PubMed: 1549105]
- 12.
- Farruggio D, Chaudhuri J, Maitra U. et al. The A1 × U72 base pair conserved in eukaryotic initiator tRNAs is important specifically for binding to the eukaryotic translation initiation factor eIF2. Mol Cell Biol. 1996;16:4248–4256. [PMC free article: PMC231423] [PubMed: 8754825]
- 13.
- Sprinzl M, Horn C, Brown M. et al. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. [PMC free article: PMC147216] [PubMed: 9399820]
- 14.
- Drabkin HJ, Helk B, RajBhandary UL. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J Biol Chem. 1993;268:25221–25228. [PubMed: 8227087]
- 15.
- Astrom SU, von Pawel-Rammingen U, Bystrom AS. The yeast initiator tRNAMet can act as an elongator tRNA(Met) in vivo. J Mol Biol. 1993;233:43–58. [PubMed: 8377191]
- 16.
- Basavappa R, Sigler PB. The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. [PMC free article: PMC453028] [PubMed: 1915284]
- 17.
- Astrom SU, Bystrom AS. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–546. [PubMed: 7954819]
- 18.
- Kiesewetter S, Ott G, Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNA(iMet) from yeast and wheat germ. Nucleic Acids Res. 1990;18:4677–4682. [PMC free article: PMC331916] [PubMed: 2395634]
- 19.
- Forster C, Chakraburtty K, Sprinzl M. Discrimination between initiation and elongation of protein biosynthesis in yeast: identity assured by a nucleotide modification in the initiator tRNA. Nucleic Acids Res. 1993;21:5679–5683. [PMC free article: PMC310535] [PubMed: 8284215]
- 20.
- Astrom SU, Nordlund ME, Erickson FL. et al. Genetic interactions between a null allele of the RIT1 gene encoding an initiator tRNA-specific modification enzyme and genes encoding translation factors in Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:967–976. [PubMed: 10485288]
- 21.
- Drabkin HJ, Estrella M, Rajbhandary UL. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol Cell Biol. 1998;18:1459–1466. [PMC free article: PMC108860] [PubMed: 9488462]
- 22.
- Donahue TF. Genetic approaches to translation initiation in Saccharomyces cerevisiaeIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000487–502.
- 23.
- Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J. 1996;15:6311–6320. [PMC free article: PMC452454] [PubMed: 8947054]
- 24.
- Huang HK, Yoon H, Hannig EM. et al. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. [PMC free article: PMC316512] [PubMed: 9308967]
- 25.
- Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. [PMC free article: PMC125960] [PubMed: 11927566]
- 26.
- Drabkin HJ, RajBhandary UL. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol Cell Biol. 1998;18:5140–5147. [PMC free article: PMC109099] [PubMed: 9710598]
- 27.
- Erickson FL, Harding LD, Dorris DR. et al. Functional analysis of homologs of translation initiation factor 2gamma in yeast. Mol Gen Genet. 1997;253:711–719. [PubMed: 9079882]
- 28.
- Laurino JP, Thompson GM, Pacheco E. et al. The beta subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol Cell Biol. 1999;19:173–181. [PMC free article: PMC83876] [PubMed: 9858542]
- 29.
- Das S, Maiti T, Das K. et al. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the beta -subunit of eIF2. J Biol Chem. 1997;272:31712–31718. [PubMed: 9395514]
- 30.
- Asano K, Krishnamoorthy T, Phan L. et al. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsi;, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. [PMC free article: PMC1171254] [PubMed: 10075937]
- 31.
- Cho S, Hoffman DW. Structure of the beta Subunit of Translation Initiation factor 2 from the Archaeon Methanococcus jannaschii: A Representative of the eIF2beta/eIF5 Family of Proteins. Biochemistry. 2002;41:5730–5742. [PubMed: 11980477]
- 32.
- Thompson GM, Pacheco E, Melo EO. et al. Conserved sequences in the beta subunit of archaeal and eukaryal translation initiation factor 2 (eIF2), absent from eIF5, mediate interaction with eIF2gamma. Biochem J. 2000;347 Pt 3:703–709. [PMC free article: PMC1221006] [PubMed: 10769173]
- 33.
- Erickson FL, Nika J, Rippel S. et al. Minimum requirements for the function of eukaryotic translation initiation factor 2. Genetics. 2001;158:123–132. [PMC free article: PMC1461651] [PubMed: 11333223]
- 34.
- Nika J, Rippel S, Hannig EM. Biochemical analysis of the eIF2beta gamma complex reveals a structural function for eIF2α in catalyzed nucleotide exchange. J Biol Chem. 2001;276:1051–1056. [PubMed: 11042214]
- 35.
- Krishnamoorthy T, Pavitt GD, Zhang F. et al. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol. 2001;21:5018–5030. [PMC free article: PMC87228] [PubMed: 11438658]
- 36.
- Nonato MC, Widom J, Clardy J. Crystal structure of the N-terminal segment of human eukaryotic translation initiation factor 2alpha. J Biol Chem. 2002;277:17057–17061. [PubMed: 11859078]
- 37.
- Sette M, van Tilborg P, Spurio R. et al. The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J. 1997;16:1436–1443. [PMC free article: PMC1169740] [PubMed: 9135158]
- 38.
- Battiste JL, Pestova TV, Hellen CUT. et al. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–119. [PubMed: 10678173]
- 39.
- Trachsel H. Binding of initiator methionyl-tRNA to ribosomesIn: Hershey JWB, Mathews MB, Sonenberg N, eds.Translational ControlCold Spring Harbor: Cold Spring Harbor Laboratory Press,1996113–138.
- 40.
- Pavitt GD, Ramaiah KV, Kimball SR. et al. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. [PMC free article: PMC316533] [PubMed: 9472020]
- 41.
- Fabian JR, Kimball SR, Heinzinger NK. et al. Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J Biol Chem. 1997;272:12359–12365. [PubMed: 9139680]
- 42.
- Gomez E, Pavitt GD. Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bϵ) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol Cell Biol. 2000;20:3965–3976. [PMC free article: PMC85753] [PubMed: 10805739]
- 43.
- Kimball SR, Fabian JR, Pavitt GD. et al. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α. Role of the alpha- and delta-subunits of eIF2B. J Biol Chem. 1998;273:12841–12845. [PubMed: 9582312]
- 44.
- Leegwater PA, Vermeulen G, Konst AA. et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–388. [PubMed: 11704758]
- 45.
- van der Knaap MS, Leegwater PA, Konst AA. et al. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–270. [PubMed: 11835386]
- 46.
- Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J Biol Chem. 1999;274:17975–17980. [PubMed: 10364246]
- 47.
- Thompson HA, Sadnik I, Scheinbuks J. et al. Studies on native ribosomal subunits from rat liver. Purification and characterization of a ribosome dissociation factor. Biochemistry. 1977;16:2221–2230. [PubMed: 861207]
- 48.
- Phan L, Zhang X, Asano K. et al. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. [PMC free article: PMC109077] [PubMed: 9671501]
- 49.
- Asano K, Phan L, Anderson J. et al. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. [PubMed: 9660829]
- 50.
- Bandyopadhyay A, Maitra U. Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucleic Acids Res. 1999;27:1331–1337. [PMC free article: PMC148320] [PubMed: 9973622]
- 51.
- Asano K, Clayton J, Shalev A. et al. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. [PMC free article: PMC316978] [PubMed: 11018020]
- 52.
- Fletcher CM, Pestova TV, Hellen CU. et al. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2637. [PMC free article: PMC1171342] [PubMed: 10228174]
- 53.
- Valasek L, Phan L, Schoenfeld LW. et al. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. [PMC free article: PMC145407] [PubMed: 11179233]
- 54.
- Block KL, Vornlocher HP, Hershey JW. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J Biol Chem. 1998;273:31901–31908. [PubMed: 9822659]
- 55.
- Phan L, Schoenfeld LW, Valasek L. et al. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 2001;20:2954–2965. [PMC free article: PMC125478] [PubMed: 11387228]
- 56.
- Lamphear BJ, Kirchweger R, Skern T. et al. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. [PubMed: 7665619]
- 57.
- Gradi A, Imataka H, Svitkin YV. et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. [PMC free article: PMC121501] [PubMed: 9418880]
- 58.
- Asano K, Shalev A, Phan L. et al. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. [PMC free article: PMC125443] [PubMed: 11331597]
- 59.
- Methot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. [PMC free article: PMC231531] [PubMed: 8816444]
- 60.
- Vornlocher HP, Hanachi P, Ribeiro S. et al. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem. 1999;274:16802–16812. [PubMed: 10358023]
- 61.
- Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. [PubMed: 1017010]
- 62.
- Varani G. A cap for all occasions. Structure. 1997;5:855–858. [PubMed: 9261078]
- 63.
- Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. [PubMed: 10872469]
- 64.
- Marcotrigiano J, Gingras A-C, Sonenberg N. et al. Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP Cell 199789951–961. 295 Mechanism of Translation Initiation in Eukaryotes . [PubMed: 9200613]
- 65.
- Matsuo H, Li H, McGuire AM. et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. [PubMed: 9302999]
- 66.
- Aravind L, Koonin EV. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–1184. [PMC free article: PMC310937] [PubMed: 10958635]
- 67.
- Raught B, Gingras A-C, Sonenberg N. Regulation of ribosomal recruitment in eukaryotesIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor, NY: Cold Spring Harbor Laboratory Press;2000245–293.
- 68.
- Tomoo K, Shen X, Okabe K. et al. Crystal structures of 7-methylguanosine 5prime prime or minute-triphosphate (m7GTP)- and P1-7-methylguanosine-P3-adenosine-5prime prime or minute,5prime prime or minute-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J. 2002;362:539–544. [PMC free article: PMC1222416] [PubMed: 11879179]
- 69.
- Minich WB, Balasta ML, Goss DJ. et al. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA. 1994;91:7668–7672. [PMC free article: PMC44463] [PubMed: 8052640]
- 70.
- Scheper GC, van Kollenburg B, Hu J. et al. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem. 2002;277:3303–3309. [PubMed: 11723111]
- 71.
- McKendrick L, Morley SJ, Pain VM. et al. Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur J Biochem. 2001;268:5375–5385. [PubMed: 11606200]
- 72.
- Lachance PE, Miron M, Raught B. et al. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol Cell Biol. 2002;22:1656–1663. [PMC free article: PMC135594] [PubMed: 11865045]
- 73.
- Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. [PMC free article: PMC1169795] [PubMed: 9155018]
- 74.
- Waskiewicz AJ, Flynn A, Proud CG. et al. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. [PMC free article: PMC1169794] [PubMed: 9155017]
- 75.
- Pyronnet S, Imataka H, Gingras AC. et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. [PMC free article: PMC1171121] [PubMed: 9878069]
- 76.
- Hentze MW. eIF4G: a multipurpose ribosome adapterγ Science. 1997;275:500–501. [PubMed: 9019810]
- 77.
- Dever TE. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. [PubMed: 10500305]
- 78.
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. [PMC free article: PMC232551] [PubMed: 9372926]
- 79.
- Dominguez D, Kislig E, Altmann M. et al. Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem J. 2001;355:223–230. [PMC free article: PMC1221730] [PubMed: 11256967]
- 80.
- Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. [PMC free article: PMC231690] [PubMed: 8943342]
- 81.
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. [PMC free article: PMC86078] [PubMed: 10913184]
- 82.
- Marcotrigiano J, Lomakin IB, Sonenberg N. et al. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. [PubMed: 11172724]
- 83.
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. [PubMed: 7550332]
- 84.
- Morino S, Imataka H, Svitkin YV. et al. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. [PMC free article: PMC85104] [PubMed: 10611225]
- 85.
- De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5' end-dependent. RNA. 1998;4:828–836. [PMC free article: PMC1369662] [PubMed: 9671055]
- 86.
- De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. [PMC free article: PMC1171558] [PubMed: 10469664]
- 87.
- Tarun SZ Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article: PMC452544] [PubMed: 9003792]
- 88.
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. [PMC free article: PMC1171091] [PubMed: 9857202]
- 89.
- Jacobson A. Poly(A) metabolism and translation: the closed-loop modelIn: Hershey JWB, Mathews MB, Sonenberg N, eds.Translational ControlCold Spring Harbor: Cold Spring Harbor Laboratory Press,1996451–480.
- 90.
- Sachs AB. Physical and functional interactions between the mRNA cap structure and the poly(A) tailIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational control of gene expressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000447–465.
- 91.
- Korneeva NL, Lamphear BJ, Hennigan FL. et al. Characterization of the two eIF4A-binding sites on human eIF4G-1. J Biol Chem. 2001;276:2872–2879. [PubMed: 11060291]
- 92.
- Li W, Belsham GJ, Proud CG. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J Biol Chem. 2001;276:29111–29115. [PubMed: 11408474]
- 93.
- Koonin EV. Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 1995;4:1608–1617. [PMC free article: PMC2143190] [PubMed: 8520487]
- 94.
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. [PubMed: 10322435]
- 95.
- Linder P, Slonimski PP. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci USA. 1989;86:2286–2290. [PMC free article: PMC286897] [PubMed: 2648398]
- 96.
- Li Q, Imataka H, Morino S. et al. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. [PMC free article: PMC84727] [PubMed: 10523622]
- 97.
- Prat A, Schmid SR, Buser P. et al. Expression of translation initiation factor 4A from yeast and mouse in Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1050:140–145. [PubMed: 2119809]
- 98.
- Rozen F, Edery I, Meerovitch K. et al. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. [PMC free article: PMC360981] [PubMed: 2304461]
- 99.
- Rogers GW Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. [PubMed: 10212190]
- 100.
- Rogers GW Jr, Richter NJ, Lima WF. et al. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. [PubMed: 11418588]
- 101.
- Pause A, Methot N, Svitkin Y. et al. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. [PMC free article: PMC394930] [PubMed: 8131750]
- 102.
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. [PubMed: 9485365]
- 103.
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. [PubMed: 9485364]
- 104.
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. [PMC free article: PMC231689] [PubMed: 8943341]
- 105.
- Coppolecchia R, Buser P, Stotz A. et al. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 1993;12:4005–4011. [PMC free article: PMC413683] [PubMed: 8404866]
- 106.
- Altmann M, Muller PP, Wittmer B. et al. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. [PMC free article: PMC413682] [PubMed: 8404865]
- 107.
- Methot N, Pause A, Hershey JW. et al. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol. 1994;14:2307–2316. [PMC free article: PMC358597] [PubMed: 8139536]
- 108.
- Naranda T, Strong WB, Menaya J. et al. Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J Biol Chem. 1994;269:14465–14472. [PubMed: 8182051]
- 109.
- Altmann M, Wittmer B, Methot N. et al. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 1995;14:3820–3827. [PMC free article: PMC394456] [PubMed: 7543843]
- 110.
- Methot N, Pickett G, Keene JD. et al. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article: PMC1369349] [PubMed: 8846295]
- 111.
- Richter-Cook NJ, Dever TE, Hensold JO. et al. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. [PubMed: 9516461]
- 112.
- Richter NJ, Rogers GW Jr, Hensold JO. et al. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem. 1999;274:35415–35424. [PubMed: 10585411]
- 113.
- Feng P, Everly DN Jr, Read GS. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J Virol. 2001;75:10272–10280. [PMC free article: PMC114601] [PubMed: 11581395]
- 114.
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. [PMC free article: PMC2115416] [PubMed: 2645293]
- 115.
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. [PubMed: 10395892]
- 116.
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. [PubMed: 2982496]
- 117.
- Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. [PubMed: 9732867]
- 118.
- Pestova TV, Hellen CU. Ribosome recruitment and scanning: what's newγ Trends Biochem Sci. 1999;24:85–87. [PubMed: 10203752]
- 119.
- Yoon HJ, Donahue TF. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992;12:248–260. [PMC free article: PMC364089] [PubMed: 1729602]
- 120.
- Naranda T, MacMillan SE, Donahue TF. et al. SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2307–2313. [PMC free article: PMC231218] [PubMed: 8628297]
- 121.
- Chaudhuri J, Si K, Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J Biol Chem. 1997;272:7883–7891. [PubMed: 9065455]
- 122.
- Choi SK, Olsen DS, Roll-Mecak A. et al. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol. 2000;20:7183–7191. [PMC free article: PMC86272] [PubMed: 10982835]
- 123.
- Carter AP, Clemons WM Jr, Brodersen DE. et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. [PubMed: 11228145]
- 124.
- Roll-Mecak A, Shin BS, Dever TE. et al. Engaging the ribosome: universal IFs of translation. Trends Biochem Sci. 2001;26:705–709. [PubMed: 11738593]
- 125.
- Pestova TV, Dever TE, Hellen CU. Ribosomal subunit joiningIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press;2000425–445.
- 126.
- Choi SK, Lee JH, Zoll WL. et al. Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. [PubMed: 9624054]
- 127.
- Lee JH, Choi SK, Roll-Mecak A. et al. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. PNAS. 1999;96:4342–4347. [PMC free article: PMC16334] [PubMed: 10200264]
- 128.
- Wilson SA, Sieiro-Vazquez C, Edwards NJ. et al. Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem J. 1999;342( Pt 1):97–103. [PMC free article: PMC1220441] [PubMed: 10432305]
- 129.
- Carrera P, Johnstone O, Nakamura A. et al. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. [PubMed: 10678180]
- 130.
- Pestova TV, Lomakin IB, Lee JH. et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. [PubMed: 10659855]
- 131.
- Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. [PMC free article: PMC1171733] [PubMed: 10581244]
- 132.
- Roll-Mecak A, Cao C, Dever TE. et al. X-Ray structures of the universal translation initiation factor IF2/ eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. [PubMed: 11114334]
- 133.
- Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational controlIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,20001–31.
- 134.
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. [PubMed: 11909525]
- 135.
- Sonenberg N, Hershey JWB, Mathews MB.eds.Translational control of gene expressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000 . [PMC free article: PMC149136]
- 136.
- Ron D, Harding HP. PERK and translational control by stress in the endoplasmic reticulumIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000547–560.
- 137.
- Chen J-J. Heme-regulated eIF2α kinaseIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000529–546.
- 138.
- Kaufman RJ. The double-stranded RNA-activated protein kinase PKRIn: Sonenberg N, Hershey JWB, Mathews MB, eds.Translational Control of Gene ExpressionCold Spring Harbor: Cold Spring Harbor Laboratory Press,2000503–527.
- 139.
- Clemens MJ. Initiation factor eIF2α phosphorylation in stress responses and apoptosis. Prog Mol Subcell Biol. 2001;27:57–89. [PubMed: 11575161]
- 140.
- Lu L, Han AP, Chen JJ. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol. 2001;21:7971–7980. [PMC free article: PMC99965] [PubMed: 11689689]
- 141.
- Harding HP, Novoa II, Zhang Y. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. [PubMed: 11106749]
- 142.
- Poulin F, Gingras A-C, Olsen H. et al. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. [PubMed: 9593750]
- 143.
- Pause A, Belsham GJ, Gingras A-C. et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. [PubMed: 7935836]
- 144.
- Gingras A-C, Raught B, Gygi SP. et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. [PMC free article: PMC312813] [PubMed: 11691836]
- 145.
- Gingras A-C, Gygi SP, Raught B. et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. [PMC free article: PMC316780] [PubMed: 10364159]
- 146.
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. [PubMed: 11297505]
- 147.
- Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. [PubMed: 10072357]
- 148.
- Dennis PB, Jaeschke A, Saitoh M. et al. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. [PubMed: 11691993]
- 149.
- Shibahara K, Asano M, Ishida Y. et al. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene. 1995;166:297–301. [PubMed: 8543179]
- 150.
- Imataka H, Olsen HS, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. [PMC free article: PMC1169682] [PubMed: 9049310]
- 151.
- Craig AW, Haghighat A, Yu AT. et al. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. [PubMed: 9548260]
- 152.
- Ishigaki Y, Li X, Serin G. et al. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. [PubMed: 11551508]
- 153.
- Ponting CP. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci. 2000;25:423–426. [PubMed: 10973054]
Publication Details
Author Information and Affiliations
Authors
Francis Poulin and Nahum Sonenberg.Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Poulin F, Sonenberg N. Mechanism of Translation Initiation in Eukaryotes. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.