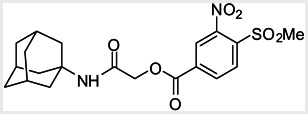
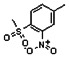
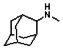
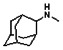
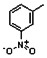
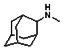
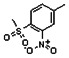
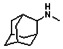
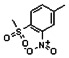
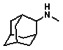
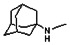
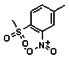
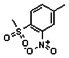
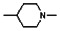
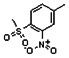
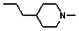
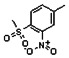
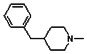
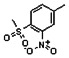
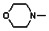
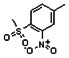
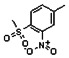
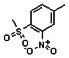
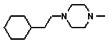
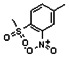
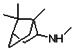
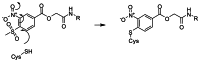Thyroid hormone receptors (TRs) are members of the nuclear hormone receptor superfamily and regulate many homeostatic processes, including basal metabolism, cardiovascular function, body weight, and lipid trafficking. Upon binding of the ligand triiodothyronine (T3), TR undergoes a conformational change that releases corepressors and recruits coactivators, such as Steroid Receptor Coactivator 2 (SRC2); in turn, these modulate the expression of target genes. In this report, we used a TRβ-SRC2 fluorescence polarization assay to screen the Molecular Libraries Small Molecule Repository (MLSMR) and identify a novel methylsulfonylnitrobenzoate (MSNB)-containing series that blocks the association of TRβ with a SRC2 peptide. This inhibitor probe molecule, ML151 (CID 5184800), blocked TRβ-SRC2 interaction with a potency of 1.8μM. Mechanistic studies revealed that ML151 (CID 5184800) is a covalent inhibitor and binds irreversibly to Cys298 within the AF-2 cleft of TRβ. This series will be useful for in vitro mechanistic studies of TR-SRC2 interactions, as well as other nuclear hormone receptor-coactivator interactions.
Assigned Assay Grant #: DK058080
Screening Center Name & PI: NIH Chemical Genomics Center, Dr. Christopher P. Austin
Chemistry Center Name & PI: NIH Chemical Genomics Center, Dr. Christopher P. Austin
Assay Submitter & Institution: Dr. Kip Guy, St. Jude Children’s Research Hospital
PubChem Summary Bioassay Identifier (AID): 2512
Recommendations for scientific use of the probe
Thyroid receptor (TR) regulates many homeostatic processes including basal metabolism, cardiovascular function, body weight, and lipid trafficking. TR modulators are potential therapeutics for obesity and hyperlipidemias, but current thyroid analogs have undesirable side effects, particularly cardiac stimulation. This probe is a member of a series that inhibits the interaction between the ligand-occupied TR and its coactivator, steroid receptor coregulator 2 (SRC2). This series can be used in vitro to study the mechanisms by which coactivators induce nuclear hormone receptor (NHR) activity in general, and how SRC2 activates TR in particular. Because SRC2 interacts with a number of different NHRs, including the androgen receptor (AR) and glucocorticoid receptor, TR selective inhibitors can help elucidate how SRC2 discriminates among different NHRs.
1. Introduction
Thyroid hormone is instrumental in controlling aspects of metabolism, growth, and development (reviewed in [1]). The thyroid receptor (TR) is a member of the family of nuclear hormone receptors (NHR) that induce the expression of transcriptional targets. There are two TR genes, TR alpha and TR beta, which mediate distinct functions. TR alpha regulates key aspects of heart function, while TR beta controls plasma cholesterol levels, as well as feedback regulation between the hypothalamus, pituitary, and thyroid. Like other NHRs, TR is composed of three parts, a transcriptional activation domain, a DNA binding domain, and a ligand binding domain. Ligand binding to TR causes the dissociation of corepressors and the subsequent association of coactivators that activate target gene expression. One coactivator that interacts strongly with TR, as well as other NHRs, is sterol receptor coactivator 2 (SRC2). SRC2 contains a nuclear receptor interaction domain composed of three short leucine-rich motifs that bind to TR. SRC2-2, the second repeat of this domain, binds to TR with an affinity comparable to full length SRC2. The goal of this project is to identify small molecule inhibitors that are TR selective by disrupting the association of SRC2.
2. Materials and Methods
All commercially available reagents and solvents were purchased and used without further purification. All microwave reactions were carried out in a sealed microwave vial equipped with a magnetic stir bar and heated in a Biotage Initiator Microwave Synthesizer. HPLC purification was performed using a Waters semi-preparative HPLC equipped with a Phenomenex Luna® C18 reverse phase (5 micron, 30× 75mm) column having a flow rate of 45 ml/min. The mobile phase was a mixture of acetonitrile and H2O, each containing 0.1% trifluoroacetic acid. During purification, a gradient of 30% to 80% acetonitrile over 8 minutes was used with fraction collection triggered by UV detection (220nM). 1H spectra were recorded using an Inova 400 MHz spectrometer (Varian). Two LCMS methods were used to analyze samples’ purity.
Method 1: Agilent 1200 series LC/MS equipped with a Zorbax™ Eclipse XDB-C18 reverse phase (5 micron, 4.6× 150mm) column having a flow rate of 1.1 ml/min. The mobile phase was a mixture of acetonitrile and H2O each containing 0.05% trifluoroacetic acid. A gradient of 5% to 100% acetonitrile over 8 minutes was used during analytical analysis.
Method 2: Acquity HPLC equipped with a Waters BEH C18, 1.7 micron, 2.1 × 50 mm column; Column Temperature: 45 degrees C; Flow: 0.5ml/min; Solvent A: 0.05% TFA in Water; Solvent B: 0.025% TFA in Acetonitrile; Gradient: 2% to 100% Solvent B over 1.3 minutes; Run Time - 3 min.
2.1. Assays
qHTS for Inhibitors of the Interaction of Thyroid Hormone Receptor and Steroid Receptor Coregulator 2. [AID 1469]
To identify inhibitors that specifically prevent the interaction of TR with the steroid receptor coregulator 2 (SRC2), a fluorescence polarization assay was screened. This assay detects interaction of the ligand-binding domain of human TR beta (TRβ) with a Texas Red labeled SRC2 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain [2]. Small molecule inhibitors that block the interaction of TR and SRC2 are detected by a decrease in fluorescence polarization.
For screening, 5 μl/well 0.6μM TRβ and 20nM SRC2 Texas Red in protein buffer (20mM Tris hydrochloride, 100mM NaCl, 10% glycerol, 1mM EDTA, 0.01% NP-40, 1mM DTT, 1μM T3 and 5% DMSO) were dispensed into black solid 1536-well plates (Grenier) using a solenoid-based dispenser. Following transfer of 23nl compound or DMSO vehicle by a pin tool, the plates were centrifuged 15 s at 1000 RPM and incubated 5 hr at ambient temperature. The plates were read by an Envision (Perkin Elmer) to detect fluorescence polarization of SRC2 Texas Red (555 nm excitation and 632 nm emission). Data were normalized to un-bound (all components except TRβ) and bound SRC2 Texas Red controls.
Table 1
Final 1536-well assay protocol.
Confirmatory assay
The confirmation and orthogonal TR assays were performed as follows, with the only difference being the SRC2-2 fluoroprobe; the confirmation assay used a Texas Red label and the orthogonal assay used a fluorescein label. Twenty μl/well 0.6μM TRβ and 20nM SRC2-2 fluoroprobe in protein buffer (20mM Tris hydrochloride, pH 7.4, 100mM NaCl, 10% glycerol, 1mM EDTA, 0.01% NP-40, 1mM DTT, 1μM T3 and 4% DMSO) was dispensed into black solid 384-well plates (Costar 3710) using a Biomek FX (Beckman Coulter) liquid handling system. Following transfer of 260nl compound (ranging from 7nM to 130μM), the plates were incubated 3 hr at ambient temperature. The plates were read by an Envision (Perkin Elmer) to detect fluorescence polarization of SRC2 Texas Red (555 nm excitation and 632 nm emission) or SRC2 Fluorescein (480 nm excitation and 535 nm emission).
Anti-target assay(s)
The androgen receptor (AR) assay (AID 2448) detects the interaction of the ligand-binding domain of human AR with a Texas Red labeled SRC2-3 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain. Small molecule inhibitors that block the interaction of AR and SRC2-3 are detected by a decrease in fluorescence polarization. Twenty μl/well 1μM liganded AR-LBD and 10nM Tx-SRC2-3 peptide in buffer (50mM HEPES, 150mM Li2SO4, 0.2mM TCEP, 10% glycerol, pH 7.2, and 4% DMSO) were dispensed into black solid 384-well plates (Costar 3710) using a Biomek FX (Beckman Coulter) liquid handling system. Following transfer of 260nl compound (ranging from 7nM to 130μM), the plates were incubated 3 hr at ambient temperature. The plates were read by an Envision (Perkin Elmer) to detect Texas Red polarization of Tx-SRC2-3 (555 nm excitation and 632 nm emission).
The vitamin D receptor (VDR) assay (AID 2455) detects the interaction of the ligand-binding domain of human VDR with an Alexa Fluor 647 labeled SRC2-3 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain. Small molecule inhibitors that block the interaction of VDR and SRC2-3 are detected by a decrease in fluorescence polarization. Twenty μl/well 1μM VDR and 5nM Alexa Fluor 647 SRC2-3 peptide in buffer (25mM PIPES, pH 6.75, 50mM NaCl, 0.01% NP-40, 6μM LG190178, and 4% DMSO) was dispensed into black solid 384-well plates (Costar 3710) using a Biomek FX (Beckman Coulter) liquid handling system. Following transfer of 260nl compound (ranging from 7nM to 130μM), the plates were incubated 3 hr at ambient temperature. The plates were read by an Envision (Perkin Elmer) to detect fluorescence polarization of Alexa Fluor 647 (620 nm excitation and 688 nm emission).
The peroxisome proliferator-activated receptor gamma (PPAR gamma) assay (AID 2449) detects the interaction of the ligand-binding domain of human PPAR gamma with a Texas Red labeled DRIP-2 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain. Small molecule inhibitors that block the interaction of PPAR and DRIP-2 are detected by a decrease in fluorescence polarization. Twenty μl/well 2μM PPAR gamma and 10nM Tx-DRIP-2 peptide in protein buffer (20mM TRIS, pH 7.5, 100mM NaCl, 0.01% NP-40, 20μM roziglitazone, and 4% DMSO) was dispensed into black solid 384-well plates (Costar 3710) using a Biomek FX (Beckman Coulter) liquid handling system. Following transfer of 260nl compound (ranging from 7nM to 130μM), the plates were incubated 3 hr at ambient temperature. The plates were read by an Envision (Perkin Elmer) to detect Texas Red polarization of Tx-DRIP-2 (555 nm excitation and 632 nm emission).
2.2. Probe Chemical Characterization
Structural verification and purity quantification were performed by 1H NMR analysis using a Varian spectrometer and by LC-MS analysis using an Agilent system in the following conditions:
Column: 3x 75 mm Luna C18, 3 micron
Run time: 4.5 min (short); 8.5 min (long)
Gradient: 4 % to 100 %
Mobile phase: Acetonitrile (0.025 % TFA), water (0.05 % TFA).
Flow rate: 0.8 to 1.0ml
Column temperature: 50 °C
Detectors: UV (220 nm, 254 nm) and MS (ESI+).
Both NMR and LC-MS analyses showed purity greater than 98% for ML151 (NCGC00188612, CID 5184800, SID 87550851). 1H NMR (400 MHz, CDCl3) δ ppm 1.65 – 1.74 (m, 6 H), 1.98 – 2.06 (m, 6 H), 2.07 – 2.14 (m, 3 H), 3.46 (s, 3 H), 4.76 (s, 2 H), 5.44 (br. s., 1 H), 8.33 (d, J=8.0 Hz, 1 H), 8.44 (dd, J=8.1, 1.5 Hz, 1 H), 8.48 (d, J=1.6 Hz, 1 H); HPLC: t1 (short) = 3.77 min and t2 (long) = 6.36 min, UV220 > 98%, UV254 > 98%; HRMS (ESI): m/z calcd for C20H24N2O7S [M+1]+ 437.1394, found 437.1394.
This compound is commercially available from Enamine (ID: T5217128).
Enamine Ltd, USA Office
Princton Corporate Plaza
7 Dear Park Drive, Suite M-3
Monmouth Junction, NJ 08852
Phone: +732 274 9150
Fax: +732 274 9151
The compound, ML151, is soluble at 10mM in DMSO. The compound is not fluorescent with blue excitation wavelengths (~340 nm). Solubility in PBS buffer: 2.9μM.
2.3. Probe Preparation
Schemes 1–3 illustrate the general synthetic strategy for the synthesis of methylsulfonylnitrobenzoate (MSNB). Methylsulfonylnitrobenzoates 5 were prepared by a two-step, one-pot procedure as described in Scheme 1. 2 – Chloroacetic chloride (2) was treated with amines 1 to give amides 3. Following this transformation, the amides reacted with acid 4 to give NSB 5. Alternatively, methylsulfonylnitrobenzoates 5 were prepared by a reaction sequence described in Scheme 2. Reaction of acid 4 and bromide 6 followed by Boc-deprotection gave acid 7, which reacted with amines 1 to generate the desired NSB 5 in high yields. The synthesis of NSB 12a and 12b is shown in Scheme 3. Reaction of 2-adamantylamine (8) and 9 gave chlorides 10. The chlorides were converted to alcohols 11 by reacting with sodium acetate followed by a hydrolysis reaction. Mitsunobu reaction of alcohols 11 and acid 4 produced 12a and 12b. All NSB analogs were purified by either preparative HPLC or column chromatography on silica gel.
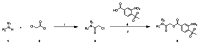
Scheme 1
Reagents and conditions: (i) iPrNEt2, ACN, r.t. 1 h; (ii) 4, MW150°C, 10 min
Scheme 2
Reagents and conditions: (i) iPrNEt2, ACN, MW150 °C, 10 min; (ii) TFA; (iii) R1R2NH, DMC, iPrNEt2

Scheme 3
Reagents and conditions: (i) iPrNEt2, DCM, r.t. 8 h; (ii) NaOAc, DMF, MW150 °C, 15 min; (iii) LiOH, THF/MeOH/H2O, MW 100 °C, 15 min; (iv) 4, DBAD, Ph3P, THF, 60 °C, 2 h
The probe molecule can be synthesized as depicted in Scheme 4. Reaction of 1-adamantylamine (14) with acid chloride 13, followed by a hydrolysis reaction gave alcohol 15. Mitsunobu reaction of the alcohol with acid 4 yielded the desired MSNB 16.
Scheme 4
Reagents and conditions: (i) iPrNEt2, DCM, r.t. o/n; (ii) LiOH, THF/MeOH/H2O, MW, 100 °C 10 min; (iii) 4, DBAD, Ph3P, THF, 60 °C, 2 h
3. Results
The titration-response data was processed by normalizing mP and total fluorescence (TF) values to controls as follows: % Activity = ((Vcompound − Vpos)/(Vpos − Vneg)) × 100, where Vcompound denotes the compound well values, Vpos denotes the median value of the DMSO-treated control wells containing TRβ, and Vneg denotes the median values of the DMSO-treated control well without TRβ (free fluoroprobe). TF was calculated as follows: TF = S + 2P where S and P are fluorescence readings in the parallel and perpendicular channels, respectively. These normalized activity values were then corrected by applying a pattern correction algorithm using DMSO-only plates placed at 24 plate intervals in the screen, as well as at the beginning and end.
Concentration response curves (CRC) were fit and classified as described [5]. Briefly, CRCs were placed into four classes. Class 1 contained complete CRCs showing both upper and lower asymptotes and r2 values > 0.9. Class 2 contained incomplete CRCs having only one asymptote and showed r2 values > 0.9. Class 3 curves were of the lowest confidence because they are poorly fit or based on activity at a single concentration point. Class 1 and 2 curves were divided further into subclasses to indicate efficacies 80% or greater (Class 1.1 and 2.1) or between 30% and 80% (Class 1.2 and 2.2). Class 4 compounds were inactives having either no curve-fit or an efficacy below threshold activity (3 SD of the mean activity). While both activators and inhibitors were recovered from the qHTS, activators were not considered further, as their activity likely arose from compound fluorescence. The qHTS resulted in 910 inhibitors (Class 1–3; Table 2), of which 511 were scored high quality (Class 1.1, 1.2 and 2.1 as well as Class 2.2 with >40% efficacy).
Table 2
Activity profile of the TR qHTS.
An in-house program was used to cluster 511 actives, yielding 730 structural series and 128 singletons. Because an active could be part of more than one series, the number of series was larger than the number of actives clustered. After clustering, structurally related compounds with inconclusive or no activity were added to each series. Each series contained at least three compounds, of which at least one was active. Series were flagged for the following potential liabilities (number of series in parentheses): concentration dependent changes in total fluorescence in the FP assay or at 547 nm excitation and 618 emission (206), promiscuous aggregation (17), promiscuous redox activity (55), low potency (> 20μM) or low efficacy (< 50%) (193), significantly lower actives compared to mean actives in all series using a Fisher’s exact test, P < 0.05 (10), and promiscuous activity in other assays (17). This process identified 8 series and two singletons with no liabilities. To prioritize compounds for follow up studies, the series were examined for previously identified scaffolds, presence of chemically undesirable functional groups, and compounds with ‘Rule of Five’ violations [6]. Confirmation studies of prioritized series were performed using the primary TR FP assay containing a Texas Red labeled fluoroprobe (AID 1573) and an orthogonal FP assay that utilized a fluorescein labeled probe (AID 2487). These tests identified the nitrosulfonyl benzoate series as the most promising for further optimization.
3.1. Summary of Screening Results
The compound library was tested using quantitative high throughput screening (qHTS), a method where each compound is tested at multiple concentrations to generate a titration-response curve for each sample [5]. The qHTS was conducted over nine days, and 291,510 samples were assayed at six concentrations. The screen performed well; of the 1418 assay plates assayed, 1380 (97%) plates passed quality control, and these showed 0.73 mean Z′ scores and 128 mean mP window. The control titration of beta-aminophenylketone inhibitor (CID 3092218) present on each plate performed consistently, showing a mean IC50 of 8.2 ± 5.2μM.
3.2. Dose Response Curves for Probe
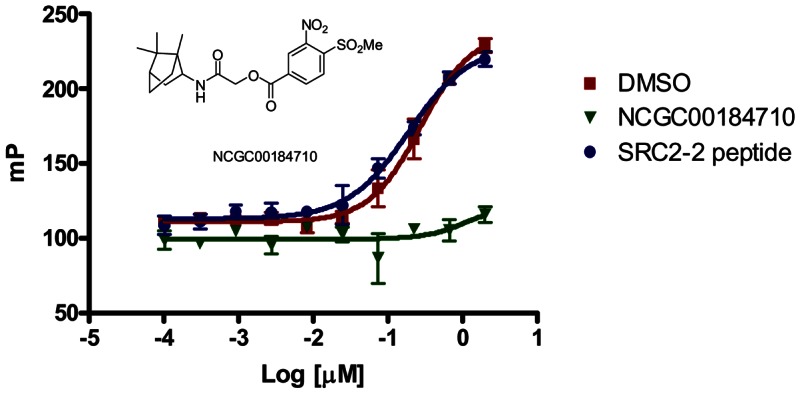
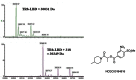
Figure 1TR inhibition by a methylsulfonylnitrobenzoate series member is irreversible
TRβ was incubated with DMSO (red square), NCGC00184710 (CID 24526448, green triangle) or SRC2-2 peptide (blue circle) for 3hr in assay buffer. Samples were then dialyzed in assay buffer overnight, recovered and tested in the TRβ FP assay at the shown concentrations of TRβ
3.3. Scaffold/Moiety Chemical Liabilities
The benzoate moiety of the probe molecule could be hydrolyzed by esterase.
3.4. SAR Tables
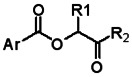
All analogs were evaluated in the confirmation TR FP assay (AID 1573). The preliminary SAR study of the aromatic (Ar), R1, and R2 positions yielded several important findings (Table 3). At the Ar position, the 3-nitro-4-sulfonyl benzoate moiety is important for inhibitory activity because both 3-nitrobenzoate 19 and 4-methylsulfonylbenzoate 18 were inactive. At the R1 position, methyl and ethyl groups had no effect on potency (compare 17, 12a and 12b). At the R2 position, hydrophilic amines such as morpholine 5e and N-methylpiperazine 5f diminished activity. Among the tested analogs, 1-adamantyl (12a, 12b, and 17–19) and 2-adamantyl 16 amines were the best groups at this position.
Table 3
Structure-activity relationships of the methylsulfonylnitrobenzoate series.
3.5. Cellular Activity
HEK293 cells were co-transfected with a CMV-TRβ expression vector, a Photonis luciferase reporter fused to a thyroid response element and a Renilla luciferase reporter, used as a control for transfection efficiency. Cells were treated with 30 nM T3 and different concentrations of ML151 (CID 5184800), incubated for 18 hours, and transcriptional reporter activities measured. ML151 (CID 5184800) showed a concentration-dependent decrease of TRβ-mediated activity over the tested concentration range of 2.5 to 20 μM (Ref 7).
4. Discussion
Small molecule inhibitors of SRC2 interaction with TR have been identified. One series, containing an aromatic beta-amino ketone core, is an irreversible inhibitor of TR and has an IC50 of 2μM [2]. Furthermore, many analogs of this series show off-target activity by blocking hERG channel activity [3]. Our goal was to identify a new chemical entity that reversibly inhibited TR and SRC2 interaction with an IC50 value less than 10μM.
Note, because the assay measures a protein-protein interaction and uses near micromolar amounts of target protein, the lower limit of potency detection is about 1μM IC50. Given the historical difficulty in achieving potent inhibitors of protein-protein interactions, the potency goals for this probe were revised to < 10μM.
The one novel series of sufficient potency identified from the screen showed irreversible binding. Thus, the probe goal was modified to identify a new chemical entity that inhibits the TR and SRC2 interaction with an IC50 less than 10μM.
4.1. Comparison to existing art and how the new probe is an improvement
This new series has several improvements over the prior art. Analogs of the aromatic beta-amino ketone series reacted to Cys298, and to a lesser degree to Lys211 and Cys388 [4], while the methylsulfonylnitrobenzoate series described here reacts only with Cys298. The presence of an amino functionality in the aromatic beta-amino ketone series has been attributed to the off target inhibition of the hERG channel [3]. The methylsulfonylnitrobenzoate series does not contain an amino group, and hERG inhibition is not expected. The aromatic beta-amino ketone series undergoes β-elimination to form a reactive intermediate that interacts with Cys298 [4]. The methylsulfonylnitrobenzoate series does not from an intermediate and can be tracked more easily than the prior aromatic beta-amino ketone series [3].
4.2. Mechanism of Action Studies
To confirm that inhibitors prevent the interaction of TRβ with the steroid receptor coregulator 2 (SRC2), a TR-SRC2 interaction assay (AID 2444) using Alpha Screen (PerkinElmer) was employed. This assay detects the interaction of the ligand-binding domain of human TRβ with a SRC2 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain. Small molecule inhibitors that block the interaction of TR and SRC2 are detected by a decrease in chemiluminescence. Fifteen μl/well of 100nM TRβ and 100nM SRC2-2-peg2-biotin in buffer (25mM HEPES, 100mM NaCl, 1mM DTT, 0.1% BSA, 0.01% NP-40) were dispensed into white 384 well Optiplates (PerkinElmer). Following transfer of 135nl compound (ranging from 90nM to 200μM), plates were incubated 1 hr, and 5μl acceptor beads (6.3 μg/ml TRβ antibody [Santa Cruz #sc-32754] and 40 μg/ml protein A beads in buffer) were added. After 30 min incubation, 5μl of streptavidin donor beads were added and after 90 min incubation, the plates were read by an Envision (Perkin Elmer) to detect luminescence of the donor and acceptor bead interaction (680 nm excitation and 520–620 nm emission). ML151 A(NCGC00188612, CID 5184800) showed 1.8μM IC50 in the TR Alpha Screen assay, which was similar to 1.4μM IC50 determined in the TR FP assay.
The aromatic β-amino ketone inhibitors of TR are irreversible and form a covalent adduct with cysteine residues in the coactivator binding pocket of TR [2]. Members of the nitrosulfonyl benzoate series were tested for irreversible binding as well. TRβ (1μM) was incubated with DMSO, SRC2-2 peptide (100μM) or NCGC00184710 (CID 24526448, 100μM) for 3 hr in assay buffer. Samples were then dialyzed in assay buffer (4l) overnight using 3000 MW cutoff Slide-A-Lyzer MINI dialysis units (Pierce, IL). The protein samples were concentrated using a 10,000 MW cutoff, spin filter column (Amicon Ultra, Millipore) and then quantified by Bradford analysis. The protein samples were serially diluted from 4 to 0.002μM in assay buffer in 96-well plates. Then, 10μl of diluted protein was added to 10μl of 40nM SRC2 fluorescence probe in 384 well plates. Unlike DMSO or SRC2-2 peptide pretreatment, NCGC00184710 (CID 24526448) pretreatment continued to inhibit SRC2-2 interaction with TRβ following dialysis (Figure 1).
We wished to identify the nature of the irreversible inhibition of TR by methylsulfonylnitrobenzoate analogs. The TRβ ligand-binding domain (LBD) was incubated in the absence or presence of NCGC00184816 (CID 42898880) for 2 hr in assay buffer, and the samples subjected to mass spectrometry. The TR LBD alone had a mass of 30,031 Dalton, but when incubated with NCGC00184816 (CID 42898880), it had a mass of 30,349 Dalton (Figure 2). The 318 Dalton difference corresponds to the size of NCGC00184816 (CID 42898880) without the methylsulfonyl group, suggesting a covalent bond was formed via an addition-elimination mechanism between one molecule of TRβ LBD and one molecule of NCGC00184816 (CID 42898880). Mass spectrometry analysis of trypsin digests of the TR-compound pair showed that NCGC00184816 (CID 42898880) was covalently attached to cysteine 298 of the protein. The proposed mechanism of action for covalent attachment of methylsulfonylnitrobenzoate analogs is shown in Figure 3. Characterization of this series in cell-based assays is ongoing.
4.3. Planned Future Studies
We plan to perform more medicinal chemistry to improve cellular activity.
5 References
- 1.
- Moore JM, Guy RK. Coregulator interactions with the thyroid hormone receptor. Mol Cell Proteomics. 2005;4(4):475–82. [PubMed: 15657066]
- 2.
- Arnold LA, Estebanez-Perpina E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. Discovery of small molecule inhibitors of the interaction of the thyroid hormone receptor with transcriptional coregulators. J Biol Chem. 2005;280(52):43048–55. [PubMed: 16263725]
- 3.
- Hwang JY, Arnold LA, Zhu F, Kosinski A, Mangano TJ, Setola V, Roth BL, Guy RK. Improvement of pharmacological properties of irreversible thyroid receptor coactivator binding inhibitors. J Med Chem. 2009;52(13):3892–901. [PMC free article: PMC2753520] [PubMed: 19469546]
- 4.
- Estebanez-Perpina E, Arnold LA, Jouravel N, Togashi M, Blethrow J, Mar E, Nguyen P, Phillips KJ, Baxter JD, Webb P, Guy RK, Fletterick RJ. Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor. Mol Endocrinol. 2007;21(12):2919–28. [PubMed: 17823305]
- 5.
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103(31):11473–8. [PMC free article: PMC1518803] [PubMed: 16864780]
- 6.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. [PubMed: 11259830]
- 7.
- Hwang JY, et al. Methylsulfonylnitrobenzoates (MSNB’s): A New Class of Irreversible Inhibitors of the Interaction of the Thyroid Hormone Receptor and its Obligate Coactivators that Functionally Antagonizes Thyroid Hormone. J. Biological Chemistry. 2011. in press. [PMC free article: PMC3069392] [PubMed: 21321127]
Publication Details
Author Information and Affiliations
Authors
Wenwei Huang,1,3 Ronald L Johnson,1 Ruili Huang,1 Jennifer Wichterman,1 Jong Yeon Hwang,2 and R Kip Guy2.3Affiliations
Publication History
Received: March 18, 2010; Last Update: March 25, 2011.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Huang W, Johnson RL, Huang R, et al. qHTS for Inhibitors of the Interaction of Thyroid Hormone Receptor and Steroid Receptor Coregulator 2. 2010 Mar 18 [Updated 2011 Mar 25]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-.