Featured Resource: Updated Entrez Sequence Database Interfaces
The Entrez sequence databases (Nucleotide, Protein, GSS, and EST) have recently completed migration to the streamlined discovery-oriented design that has been in service in PubMed for nearly a year, described fully in the November 2009 issue of the NCBI News. The sequence database re-design includes new homepages, a simpler interface, and new options for downloading, displaying sequences, and connecting to related data.
New homepages
The sequence database homepages now have a simplified design with three columns of links with access to information about using the resource, tools for submitting data, searching and analysis, and other related resources at the NCBI site. As in the PubMed and the Site Guide (NCBI Homepage), the new sequence database pages have the new header including the search bar with access to all NCBI Entrez databases and the Resources and How To pull-down lists to aid navigation and to access to practical task-oriented help and the footer that provides rapid navigation to all major areas of the NCBI site. Figure 1 shows the new Entrez nucleotide homepage.

Figure 1
The new nucleotide homepage with access to related resources. The new sequence database homepages include the NCBI header and footer (not shown) that provide easy navigation to other parts of the site and links to task-oriented help documentation.
Improvements to the search interface
The new interface is simplified eliminating the four control tabs of the previous version: Limits, Preview/Index, History, Clipboard, and Details. These functions are still available but are now easier to access and use. A redesigned Limits page is linked at the upper-right of the Search Box that appears on all pages. The new Advanced search page also linked above the Search Box combines the functions of the old Preview/Index and History tabs. The Search details providing the translation of the query by the Entrez engine is linked on the Advanced search page but is also shown in the right-hand Discovery column as in the new PubMed. The Clipboard, when populated, is now accessible as a link at the top of the Discovery column as described below.
Using the Advanced search page
Figure 2 shows the Advanced search page for Entrez protein. The page functions as an independent search interface that allows formulation of complex queries. The Search builder and Search history sections replace the previous Preview/Index page and facilitate the construction of more precise queries. The pull-down list in the Search Builder shows all of the fields indexed for a particular database. The Show Index link opens an alphabetical list of terms for the selected field. When a term is entered in the Search Builder, the index will open to the closest match in the index. The Add to Search Box button puts the field-restricted queries into the Search Box. These may be run using the Search button or may be added to the Search History using the Preview button. Entries in the Search History may be combined to give very precise results. The example in Figure 2 combines searches for frogs (#5), RefSeq proteins (#2), and prolactin (#3) to obtain the prolactin protein records for Xenopus laevis, NP_001086486, and Xenopus (Siluriana) tropicalis, NP_001093699).
Search results and other multiple record pages
Search result pages showing document summaries and all other multiple records views now incorporate fully the features of the PubMed redesign. Figure 3 shows the new search results for a protein search – prolactin[protein name] AND (Birds OR Mammals). The document summaries are presented in a new format with the record title first and hyperlinked to the record view. The summary now also shows the length of the sequence. The sequence identifiers, accession version and GI number, are listed below the summary. The old links menu has been removed from the document summaries in search results. However links to retrieve related sequences (similar by BLAST) are shown under the summary by default in the nucleotide and protein databases. The protein summaries also include a link to identical sequences. All links available for each record can be displayed if desired though individual settings in a My NCBI account. See the My NCBI Help manual for more information on customizing these preferences.
Multiple-record displays in one of the full-record formats such as GenBank or FASTA have the same set of controls in the new style as the summaries. The only additional feature is the Customize view control, described for single sequence views in the March 2009 NCBI News. In the case of multiple records, this control allows toggling the reverse complement of the displayed sequences.
New items in the right-hand Discovery column
The search filters that formerly were a series of tabs are now implemented as a set of links at the upper part of the right-hand Discovery column. In the case of the results shown in Figure 3, the RefSeq filter has been clicked filtering the output to show only the six Reference Sequences. Clicking the plus sign (+) at the right of the selected filter will add the filter as a term to the search. The Search details, previously a control tab that shows how the query is interpreted or mapped to Entrez controlled vocabularies is now exposed in the Discovery column. The Search details at the bottom right of Figure 3 show how the terms Birds and Mammals were expanded, translated, and mapped to the organism field (NCBI Taxonomy) to generate more accurate results. The former Clipboard tab now appears as a link above the right-hand column only if the NCBI Clipboard contains items (Figure 3, upper right).
Also in the Discovery column are two new items, Analyze these sequences and Find related data that provide access to analysis tools and pre-computed relationships – true Discovery components. Analyze these sequences, available for displays containing 20 or fewer records, provides direct access to the NCBI BLAST service from all sequence databases and, for multiple protein records, the NCBI multiple-alignment tool COBALT, described in the May 2009 NCBI News. This is a convenient interface to COBALT since it allows direct submission after using Entrez to collect the desired input set. For example the set of homologous prolactin proteins from mammals and birds in Figure 3 can now be easily aligned using COBALT.
The links shown in the Find related data list previously were presented on the Display pull-down list at the top of the results. The new location in the Discovery column makes these links easier to find and use. These provide access to the wealth of pre-computed and pre-compiled relationships that make Entrez a powerful discovery system. In many cases there may be more than one kind of link to another database. The inset in Figure 3 shows the multiple links to the nucleotide database available from the protein data. Selecting “Encoding mRNA” and clicking Find items links to the six corresponding RefSeq mRNA records.
Summary
The updated Entrez interface now implemented in the sequence databases provides a streamlined and less complex search interface as well as improved consistency of form and function across the molecular and literature resources making the NCBI site easier to use. New options for displaying and downloading records especially the ability to download coding regions are important improvements to the utility and flexibility of the molecular biology Web services at the NCBI. The presence of analysis tools and improved access to related data on search results and record displays increase the power of Entrez as a system for scientific discovery.
New Databases and Tools
Bibliography Management
My NCBI will replace eRA Commons for grantee bibliography management. The My Bibliography function in My NCBI will link with Commons to allow scientists to maintain and manage a list of their publications including journal articles, book chapters, meeting abstracts, talks and presentations, patents, and other materials. Journal articles can be those found in PubMed, journals not indexed or not yet appearing in PubMed, or manuscripts submitted to NIHMS. Citations may not be entered manually into eRA Commons at this point, and users must use My Bibliography to manage their bibliographies. For more information, please see the NIH Announcement.
NCBI Discovery Workshops
NCBI will present a two-day workshop on September 29-30, on the NIH campus in Bethesda, MD. The workshops provide hands-on experience exploring practical examples using tools and databases on the NCBI website. The four workshops are Sequences, Genomes, and Maps; Proteins, Domains and Structures; NCBI BLAST Services; and Human Variation and Disease Genes. For more information see the Discovery Workshop page, which also includes a registration link.
Microbial Genomes
Twenty-one finished microbial genomes were released in July 2010. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 178.0 is available through the NCBI web and FTP sites. The current release includes information available as of June 11, 2010. Release notes are available on the on the NCBI ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
Updates and Enhancements
My NCBI now offers sequence database preferences
My NCBI allows Preferences to be set for record format and result display settings for the Entrez sequence databases (Nucleotide, Protein, EST and GSS). Record formats can be changed from the default (GenBank, GenPept, EST, GSS) to FASTA or graphics for Nucleotide and Protein or to GenBank or FASTA for EST or GSS records. All available options on the Display settings menu in the sequence databases can also be changed from the default settings. The relevant preferences dialogs from My NCBI are shown below.
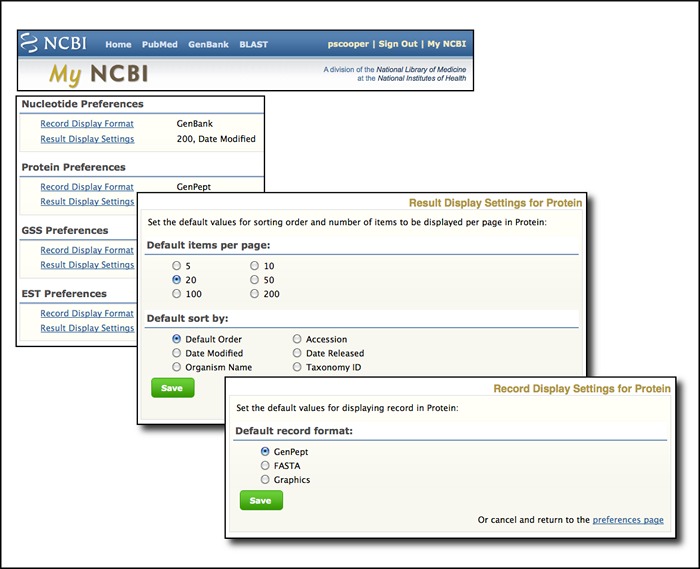
RefSeq
RefSeq Release 42 is available through the Entrez system and can be downloaded from the FTP site (ftp.ncbi.nlm.nih.gov/refseq/release). This full release incorporates genomic, transcript, and protein data available as of July 21, 2010. It includes 15,038,858 records from 10,728 different organisms. Changes since the last release can be found in the release notes (ftp.ncbi.nih.gov/refseq/release/release-notes/RefSeq-release42.txt)
New FTP file for Gene
Entrez Gene calculates matches between NCBI and Ensembl annotations and reports the matches in the Entrez gene Full Report display, the "matches Ensembl" index property, and the gene2ensembl FTP file. A new FTP file, README_ensembl, will soon be added to provide a summary of species whose annotations have been compared, including release and assembly information, and the date when the comparison was last performed. A complete description of the file is on the ftp site: ftp.ncbi.nih.gov/gene/DATA/README
Announce Lists and RSS Feeds
Eighteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html.
Twelve RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/.
Users can also stay updated on NCBI’s resources on Facebook and Twitter: twitter.com/NCBI.
Send comments and questions about NCBI resources to: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
Publication Details
Author Information and Affiliations
Publication History
Created: July 31, 2010.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Cooper P, Lipshultz D. NCBI News, July 2010. 2010 Jul 31. In: NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
![Figure 3. Entrez protein search results for the query Prolactin[protein name] AND (Birds OR Mammals) showing the new summary style.](/books/NBK45527/bin/search_summary.gif)



![Figure 3. Entrez protein search results for the query Prolactin[protein name] AND (Birds OR Mammals) showing the new summary style.](/books/NBK45527/bin/search_summary.jpg)
