The tripartite motif (TRIM) proteins harboring the RING finger, B-box and coiled-coil (RBCC) domain motifs form a large protein family. The members of this family are involved in various biological processes, including growth, differentiation, apoptosis and transcription and also in diseases and oncogenesis. Recent studies have revealed that TRIM proteins play key roles in innate antiviral immunity. An accumulating body of evidence has demonstrated that some TRIM proteins function as E3 ubiquitin ligases in specific ubiquitin-mediated protein degradation pathways; however, the precise mechanisms underlying this function have not been fully elucidated. In this review, we focus on the TRIM family of proteins specially with regard to E3 ligase.
INTRODUCTION
A large number of proteins (more than 17,000) harboring the RING finger domain have been identified in diverse eukaryotes in the simple modular architecture research tool (SMART) database. The RING finger motif is defined as a unique linear series of conserved cysteine and histidine residues, i.e., Cys-X2-Cys-X11-16-Cys-X-His-X2-Cys-X2-Cys-X7-74-Cys-X2-Cys (C3HC4 type), where X can be any amino acid. Three-dimensional analyses of RING domains have confirmed that the RING finger motif is composed of a unique "cross-brace" arrangement with 2 zinc ions and it folds into a compact domain comprising a small central β sheet and an α helix.1,2 Frequently, the RING domain is associated with cysteine-rich B-box domains followed by a predicted coiled-coil domain. This ensemble of a RING domain, 1 or 2 B-box domains and a coiled-coil domain are called RBCC or TRIM.3 Each domain of the TRIM protein may operate as a functional unit, either independently or in combination with other domains and play important roles in the recognition of specific interacting partners or the formation of oligomers. Studies have shown that the RING finger proteins play crucial roles in growth, differentiation, transcription, signal transduction and oncogenesis.4-10 These studies have revealed some essential roles played by the RING finger domains in the function of these proteins. Recently, it was revealed that the RING finger proteins are often involved in the ubiquitin-mediated protein degradation pathway.
TRIM PROTEINS ARE INVOLVED IN PROTEIN MODIFICATION PATHWAY BY UBIQUITIN
Ubiquitin-dependent protein degradation is a specific and elaborate mechanism in which the target protein to be destroyed is tagged with ubiquitin. Ubiquitination is accomplished by a complex process involving the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3) (Fig. 1).11 Ubiquitin ligase mediates the transfer of ubiquitin from E2 to a substrate, enabling degradation of the latter by the 26S proteasome. The C3HC4-type RING finger domain is found in several E3 proteins, including Cbl,12 BRCA1,13 estrogen-responsive finger protein (Efp)14 and murine double minute 2 (Mdm2) 15 and so on. The RING-H2 subtype in which Cys5 is substituted with histidine is found in RING box protein 1 (Rbx1) and anaphase promoting complex (APC) subunit 11 (Apc11), which are components of the Skp1-Cullin-F-box (SCF) and APC E3 complexes,16 respectively and in several other ubiquitin ligases. E3 enzyme is thought to be important for specific recognition of the substrate in the ubiquitination pathway. In the ubiquitination model established many years ago, the first ubiquitin moiety is anchored via its COOH-terminal Gly residue to an -NH2 group of an internal Lys residue in the target substrate. This is followed by the generation of a polyubiquitin chain in which additional ubiquitin moieties are linked to one another via Gly-76-Lys-48 isopeptide bonds.17 During recent years, other modes of ubiquitination have been discovered. One such noncanonical conjugation reaction involves the anchoring of ubiquitin to Lys residues other than Lys-48 in the previously conjugated ubiquitin moiety. The ubiquitination of Lys-63 appears to play a role in a variety of processes, including the endocytosis of cell surface receptors,18,19 postreplicative DNA repair,20 stress responses,21 ribosomal functioning,22 and activation of the IB signaling complex.23 This type of modification does not appear to involve proteolysis of the target substrates but plays a role in the activation/inactivation of the target protein.
EFP/TRIM25 FUNCTIONS AS E3 LIGASE FOR BOTH UBIQUITINATION AND ISGYLATION
In the RING finger protein family, Efp is a member of the RING-finger, B1- and B2-box, coiled coil and SPRY (RBCC-SPRY) subfamily. Efp was isolated as an estrogen-responsive gene by the genomic binding-sites cloning method utilizing a recombinant estrogen receptor (ER) protein.24 The estrogen-responsive element (ERE) to which ER can bind is found at the 3'-untranslated region (UTR) in the Efp gene and the gene's expression is predominantly detected in female reproductive organs, including the uterus, ovary and mammary gland,25 and in breast and ovarian cancers.26 Estrogen-induced Efp expression is found in the uterus, brain and mammary gland cells. Efp-knockout mice have underdeveloped uteri and estrogen responses are markedly attenuated in uteri of knockout mice; this suggests that Efp is necessary for estrogen-induced cell growth.27 Moreover, the growth of MCF7 breast cancer cells implanted in female athymic mice is reduced by treatment with the Efp antisense oligonucleotide. In contrast, Efp-overexpressing MCF7 cells in ovariectomized athymic mice generate tumors in the absence of estrogen.14 These findings indicate that Efp mediates estrogen-dependent growth in breast cancer cells. We identified 14-3-3σ , which is responsible for reduced cell growth, as a factor that interacts with Efp and detected 14-3-3σ accumulation in the embryonic fibroblasts of Efp-knockout mice. Furthermore, it has been revealed that Efp is an E3 ubiquitin ligase that targets the proteolysis of 14-3-3σ (Fig. 2). In particular, the RING domain that preferentially binds to E2 UbcH8 is essential for the ubiquitination of 14-3-3σ . The 14-3-3σ degradation after ubquitination coupled with Efp as an E3 lagase, followed by activation of the cyclin-Cdk complexes, leads to cell cycle progression, cell proliferation and tumor growth; these findings provide an insight into the cell-cycle machinery and tumorigenesis of breast cancer cells.
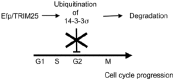
Figure 2
Model for tumor growth controlled by Efp. Efp stimulates tumor growth by targeting 14-3-3σ (a negative regulator of the cell cycle) for proteolysis as an E3 ubiquitin ligase.
In addition, we showed that Efp expression is up-regulated by Type I interferon (IFN) through an IFN-stimulated response element (ISRE) located in the first intron.28 Electrophoretic mobility shift assays and chromatin immunoprecipitation assays showed that the ISRE binds to signal transducer and activator of transcription 1 (STAT1). Moreover, we showed that the Efp protein could be conjugated with not only ubiquitin but also ISG15, a ubiquitin-like molecule.28 The IFN-stimulated gene 15 (ISG15) has been originally identified as a type I IFN-stimulated gene encoding a 15-kD protein.29 The ISG15 protein contains 2 conserved ubiquitin-like domains and is conjugated to target proteins or ISGylated along with E1, E2 and E3 by a process similar to ubiquitination (Fig. 3).30 Thus far, Efp and Herc5 have been identified as E3 ligases that undergo ISGylation.28,31,32 In particular, Efp can conjugate with both ISG15 and ubiquitin and can also conjugate these molecules to 14-3-3σ .14,28,32 ISG15 expression and protein ISGylation are strongly induced by viral infection;33,34 this finding indicates that the ISGylation system is a novel pathway that transduces the antiviral response in IFN-stimulated cells. It has been reported that ISG15 can inhibit the release of human immunodeficiency virus type 1 (HIV-1) virions35 and attenuate Sindbis virus infection.36 ISG15-/- mice are more susceptible to infection with the influenza virus, herpes simplex virus type 1 and Sindbis virus.37 These data suggest that Efp is an IFN-responsive gene that potentially mediates the functions of IFN and is involved in the ISGylation and ubiquitination of proteins, including Efp itself.
Recently, we reported that TRIM25 induces ubiquitination of retinoic acid-inducible gene I product (RIG-I), which is a cellular sensor of RNA virus infection and induces IFN-β expression (Fig. 4).38 RIG-I encodes two caspase recruitment domains (CARDs) at the N terminus and an RNA helicase domain at the C terminus. The helicase domain recognizes viral dsRNA and the CARDs directly transmits a signal that activates both IRF-3 and NFκB to induce the expression of IFN-β and the antiviral cytokine gene.39 The C-terminal SPRY domain of TRIM25 interacts with the N-terminal CARDs of RIG-I; this interaction effectively delivers the Lys-63-linked ubiquitin moiety to the CARDs of RIG-I, resulting in a marked increase in the downstream signaling activity of RIG-I. The Lys-172 residue of RIG-I is critical for efficient TRIM25-mediated ubiquitination, mitochondrial antiviral signaling protein (MAVS) binding and RIGI-mediated induction of antiviral signal transduction. TRIM25-/- mouse embryonic fibroblasts (MEFs) showed that TRIM25 is essential not only for RIG-I ubiquitination but also for RIG-I-mediated IFN-β production and antiviral activity in response to RNA virus infection.38,40 Thus, TRIM25 E3 ubiquitin ligase induces the Lys-63-linked ubiquitination of RIG-I, which is crucial for innate host antiviral immunity elicited by the RIGI signaling pathway. Moreover, the influenza A virus nonstructural protein 1 (NS1) inhibits ubiquitination of the TRIM25-mediated RIG-I CARD and RIG-I signal transduction. TRIM25 is therefore a key host factor targeted by influenza virus infection.41
E3 UBIQUITIN LIGASES IN TRIM/RBCC PROTEINS
The presence of RING finger domains in E3 ubiquitin ligases implies that the members of this TRIM family are potential targets for specific regulators/adopters in ubiquitin-dependent protein degradation. In fact, some genes belonging to the TRIM family have been shown to have E3 ligase activity. Next, we discuss such TRIM family members, focusing on the findings of recent studies.
MID1/TRIM18, MID2/TRIM1
A positional cloning approach has revealed that MID1 is involved in the Opitz GBBB syndrome (OS).42-44 Further, MID1 associates with microtubules, whereas its mutant forms do not.45 These findings suggest that MID1 plays a physiological role in microtubule dynamics. The α4 protein, a regulatory subunit of protein phosphatase 2A (PP2A)46 was isolated using yeast two-hybrid screening with MID1 as bait. The cellular localization of MID1 and α4 is coincident with cytoskeletal structures. The expression of MID1 with a mutation at the C terminus, which mimics the mutant protein in some individuals with OS, causes the formation of cytoplasmic clumps containing both proteins. A cytosolic PP2A was identified as the substrate for the E3 ligase activity of MID1. In contrast, the addition of a proteasome inhibitor to OS-derived fibroblasts expressing dysfunctional MID1 does not upregulate either PP2A or the enzyme's polyubiquitinated forms;47 this suggests that MID1 mutations result in decreased proteolysis of the C subunit of PP2A in individuals with OS.
TRIM11
TRIM11 belongs to a protein family composed of a RING finger domain, which is a putative E3 ubiquitin ligase; a B-box domain; a coiled-coil domain; and an SPRY domain. A recent experiment with yeast two-hybrid screening has revealed that TRIM11 can interact with humanin,48 which is a newly identified anti-apoptotic peptide that specifically suppresses Alzheimer's disease (AD)-related neurotoxicity. Humanin binds with Bax and prevents the translocation of Bax to mitochondria, thus preventing the release of cytochrome c.49 Thus, humanin seems to exert its anti-apoptotic effects by interfering with Bax function. The intracellular level of humanin was drastically reduced in the presence of TRIM11 and mutation of the RING finger domain or treatment with a proteasome inhibitor attenuates the effect of TRIM11 on the intracellular level of humanin.48 These results suggest that TRIM11 may act as an E3 ligase in the ubiquitin-mediated degradation of humanin.
SSA/Ro (SSA1/TRIM21)
TRIM21 has been identified as an autoantigen in Sjögren syndrome.49,50 A recent study reported that TRIM21 exhibits E3 activity and interacts with the human IgG1 heavy chain. The IgG1 heavy chain is polyubiquitinated by TRIM21 and degraded through the ubiquitin-proteasome system; this suggests that it plays a significant role in the quality control of IgG1.51 In addition, TRIM21 ubiquitinates IRF-8 and enhances IL-12p40 expression in IFN-γ/TLR-stimulated macrophages. These findings imply that TRIM21 contributes to the elicitation of innate immunity in macrophages.52
TRIM32
TRIM32 belongs to the TRIM protein family and possesses 6 C-terminal NHL domains. A point mutation in 1 NHL domain (D487N) has been linked to 2 forms of muscular dystrophy—limb girdle muscular dystrophy type 2H and sarcotubular myopathy. TRIM32 is primarily expressed in skeletal muscles and its expression is significantly elevated in muscles undergoing remodeling and during myogenic differentiation. TRIM32 ubiquitinates actin and thus probably participates in myofibrillar protein turnover, especially during muscle adaptation.53
MuRF1/TRIM63
MuRF1/TRIM63 and 2 other members of the MuRF family, namely, MuRF2/TRIM55 and MuRF3/TRIM54, all belong to the TRIM family. MuRF proteins localize to the sarcomere,54 and MuRF1 associates with titin in the M band of the sarcomere, thus possibly maintaining the stability of the sarcomeric M band.55,56 MuRF-1 is an E3 ubiquitin ligase that acts on troponin I and is upregulated in atrophic muscles.57,58 Muscular atrophy, which is associated with various diseases and is a side effect of treatment with synthetic glucocorticoid treatment, is characterized by accelerated protein degradation via the ubiquitin proteasome system. MuRF1-/- mice exhibit increased resistance to muscular atrophy and significant muscle preservation after denervation; this suggests that MuRF-1 plays a critical role in muscle turnover.58,59
TRIM5
TRIM5α, which is a member of the TRIM-SPRY subfamily, has recently been identified as a cellular factor that is essential for retroviral restriction and targets incoming retroviral capsids after viral penetration60 or the Gag assembly during HIV-1 production by rapidly degrading HIV-1 Gag polyproteins (Fig. 5).61 HIV-1 infection is blocked by rhesus monkey TRIM5α but not by human TRIM5α, whereas N-tropic murine leukemia virus (N-MLV) infection is blocked by both rhesus monkey TRIM5α and human TRIM5α.62,63 These findings imply that species-specific variations in TRIM5α govern its ability to block infection by diverse retroviruses. The RING and SPRY domains are required for this restrictive activity and variations in the latter determine the specificity of retroviral restriction.64 Indeed, the SPRY domain of TRIM5α appears to associate with the major core protein CAp30 of N-MLV but not with that of B-MLV.65 Polyubiquitylation and rapid degradation of TRIM5α require intact RING and B-box domains; however, rapid turnover of TRIM5α is not required for its antiretroviral activity.66 Our study revealed that the mRNA and protein expression of human TRIM5α is induced by IFN and that the transcription factor STAT1 is bound to the ISRE in the gene promoter.67 The human TRIM5α gene is located at the chromosomal position 11p15, clustering with other TRIM genes, namely, TRIM21, TRIM6, TRIM34 and TRIM22, which are also known as IFN-inducible genes. This suggests that TRIM5α activity could be modulated by IFN.
CONCLUSION
We have summarized the structural characteristics and functions of TRIM family proteins, specifically with regard to E3 ligase activity. However, relatively few proteins have been proven to be E3 ligases and the function of most of the members of this family remains unclear. Investigation of RING finger proteins as possible novel E3 ligases would uncover important mechanisms underlying cellular protein degradation and modification and provide new insights into the physiological and pathophysiological roles of these family members. In particular, functional analysis of the RING finger proteins involved in innate antiviral immunity can open up new fields of research. In addition to TRIM5 and TRIM25, some members of the TRIM family are known to be regulated by IFN. These proteins may play a role in viral infection by inducing posttranscriptional modifications in their target proteins. In order to understand the functions of TRIM family, it is necessary to identify the substrate of TRIM E3 ligase. We then need to clarify how the substrates are modified by ubiquitine and/or ubiquitin-like molecules and play roles in cells and in vivo.
REFERENCES
- 1.
- Borden KL, Boddy MN, Lally J, et al. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. Embo J. 1995;14(7):1532–1541. [PMC free article: PMC398240] [PubMed: 7729428]
- 2.
- Barlow PN, Luisi B, Milner A, et al. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237(2):201–211. [PubMed: 8126734]
- 3.
- Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. Embo J. 2001;20(9):2140–2151. [PMC free article: PMC125245] [PubMed: 11331580]
- 4.
- Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17(9):344–345. [PubMed: 1412709]
- 5.
- Kakizuka A, Miller WH Jr, Umesono K, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66(4):663–674. [PubMed: 1652368]
- 6.
- deThe H, Lavau C, Marchio A, et al. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66(4):675–684. [PubMed: 1652369]
- 7.
- Goddard AD, Borrow J, Freemont PS, et al. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254(5036):1371–1374. [PubMed: 1720570]
- 8.
- Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. [PubMed: 7545954]
- 9.
- Le Douarin B, Zechel C, Garnier JM, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. Embo J. 1995;14(9):2020–2033. [PMC free article: PMC398302] [PubMed: 7744009]
- 10.
- Takahashi R, Deveraux Q, Tamm I, et al. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem. 1998;273(14):7787–7790. [PubMed: 9525868]
- 11.
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. [PubMed: 9759494]
- 12.
- Joazeiro CA, Wing SS, Huang H, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286(5438):309–312. [PubMed: 10514377]
- 13.
- Lorick KL, Jensen JP, Fang S, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96(20):11364–11369. [PMC free article: PMC18039] [PubMed: 10500182]
- 14.
- Urano T, Saito T, Tsukui T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417(6891):871–875. [PubMed: 12075357]
- 15.
- Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 2001;12(4):175–186. [PubMed: 11331246]
- 16.
- Seol JH, Feldman RM, Zachariae W, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13(12):1614–1626. [PMC free article: PMC316801] [PubMed: 10385629]
- 17.
- Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11(13):1055–1066. [PubMed: 9367341]
- 18.
- Hicke L. Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9(3):107–112. [PubMed: 10201076]
- 19.
- Strous GJ, Govers R. The ubiquitin-proteasome system and endocytosis. J Cell Sci. 1999;112(10):1417–1423. [PubMed: 10212136]
- 20.
- Spence J, Sadis S, Haas AL, et al. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. [PMC free article: PMC230349] [PubMed: 7862120]
- 21.
- Arnason T, Ellison MJ. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14(12):7876–7883. [PMC free article: PMC359326] [PubMed: 7969127]
- 22.
- Spence J, Gali RR, Dittmar G, et al. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102(1):67–76. [PubMed: 10929714]
- 23.
- Deng L, Wang C, Spencer E, et al. Activation of the IkB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. [PubMed: 11057907]
- 24.
- Inoue S, Orimo A, Hosoi T, et al. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc Natl Acad Sci USA. 1993;90(23):11117–11121. [PMC free article: PMC47933] [PubMed: 8248217]
- 25.
- Orimo A, Inoue S, Ikeda K, et al. Molecular cloning, structure and expression of mouse estrogen-responsive finger protein Efp. Co-localization with estrogen receptor mRNA in target organs. J Biol Chem. 1995;270(41):24406–24413. [PubMed: 7592654]
- 26.
- Ikeda K, Orimo A, Higashi Y, et al. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000;472(1):9–13. [PubMed: 10781795]
- 27.
- Orimo A, Inoue S, Minowa O, et al. Underdeveloped uterus and reduced estrogen responsiveness in mice with disruption of the estrogen-responsive finger protein gene, which is a direct target of estrogen receptor alpha. Proc Natl Acad Sci USA. 1999;96(21):12027–12032. [PMC free article: PMC18406] [PubMed: 10518570]
- 28.
- Nakasato N, Ikeda K, Urano T, et al. A ubiquitin E3 ligase Efp is up-regulated by interferons and conjugated with ISG15. Biochem Biophys Res Commun. 2006;351(2):540–546. [PubMed: 17069755]
- 29.
- Blomstrom DC, Fahey D, Kutny R, et al. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem. 1986;261(19):8811–8816. [PubMed: 3087979]
- 30.
- Staub O. Ubiquitylation and isgylation: overlapping enzymatic cascades do the job. Sci STKE. 2004;10:pe43. [PubMed: 15304665]
- 31.
- Wong JJ, Pung YF, Sze NS, et al. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA. 2006;103(28):10735–10740. [PMC free article: PMC1484417] [PubMed: 16815975]
- 32.
- Zou W, Wang J, Zhang DE. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun. 2007;354(1):321–327. [PMC free article: PMC1858649] [PubMed: 17222803]
- 33.
- Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267(11):7806–7813. [PubMed: 1373138]
- 34.
- Ritchie KJ, Hahn CS, Kim KI, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10(12):1374–1378. [PubMed: 15531891]
- 35.
- Okumura A, Lu G, Pitha-Rowe I, et al. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103(5):1440–1445. [PMC free article: PMC1360585] [PubMed: 16434471]
- 36.
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, et al. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79(22):13974–13983. [PMC free article: PMC1280211] [PubMed: 16254333]
- 37.
- Lenschow DJ, Lai C, Frias-Staheli N, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104(4):1371–1376. [PMC free article: PMC1783119] [PubMed: 17227866]
- 38.
- Gack MU, Shin YC, Joo CH, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. [PubMed: 17392790]
- 39.
- Yoneyama M, Kikuchi M, Matsumoto K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5 and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. [PubMed: 16116171]
- 40.
- Poeck H, Inoue S, Ruland J, et al. Nat Immunol. in press; RIG-I is a dual activator of Card9 and inflammasome signaling for IL-1β production upon RNA virus recognition. [PubMed: 19915568]
- 41.
- Gack MU, Albrecht RA, Urano T, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5(5):439–449. [PMC free article: PMC2737813] [PubMed: 19454348]
- 42.
- Opitz JM. G syndrome (hypertelorism with esophageal abnormality and hypospadias, or hypospadias-dysphagia, or "Opitz-Frias" or "Opitz-G" syndrome)—perspective in 1987 and bibliography. Am J Med Genet. 1987;28(2):275–285. [PubMed: 3322001]
- 43.
- Robin NH, Opitz JM, Muenke M. Opitz G/BBB syndrome: clinical comparisons of families linked to Xp22 and 22q and a review of the literature. Am J Med Genet. 1996;62(3):305–317. [PubMed: 8882794]
- 44.
- Quaderi NA, Schweiger S, Gaudenz K, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17(3):285–291. [PubMed: 9354791]
- 45.
- Gaudenz K, Roessler E, Quaderi N, et al. Opitz G/BBB syndrome in Xp22: mutations in the MID1 gene cluster in the carboxy-terminal domain. Am J Hum Genet. 1998;63(3):703–710. [PMC free article: PMC1377398] [PubMed: 9718340]
- 46.
- Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27(3):286–291. [PubMed: 11242110]
- 47.
- Short KM, Hopwood B, Yi Z, et al. MID1 and MID2 homo- and heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, alpha 4, to microtubules: implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 2002;3(1):1. [PMC free article: PMC64779] [PubMed: 11806752]
- 48.
- Niikura T, Hashimoto Y, Tajima H, et al. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur J Neurosci. 2003;17(6):1150–1158. [PubMed: 12670303]
- 49.
- Sibilia J. Ro(SS-A) and anti-Ro(SS-A): an update. Rev Rhum Engl Ed. 1998;65(1):45–57. [PubMed: 9523386]
- 50.
- Itoh Y, Reichlin M. Autoantibodies to the Ro/SSA antigen are conformation dependent. I: Anti-60 kD antibodies are mainly directed to the native protein; anti-52 kD antibodies are mainly directed to the denatured protein. Autoimmunity. 1992;14(1):57–65. [PubMed: 1284379]
- 51.
- Takahata M, Bohgaki M, Tsukiyama T, et al. Ro52 functionally interacts with IgG1 and regulates its quality control via the ERAD system. Mol Immunol. 2008;45(7):2045–2054. [PubMed: 18022694]
- 52.
- Kong HJ, Anderson DE, Lee CH, et al. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179(1):26–30. [PubMed: 17579016]
- 53.
- Kudryashova E, Kudryashov D, Kramerova I, et al. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. [PubMed: 16243356]
- 54.
- Centner T, Yano J, Kimura E, et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001;306(4):717–726. [PubMed: 11243782]
- 55.
- Gregorio CC, Perry CN, McElhinny AS. Functional properties of the titin/connectin-associated proteins, the muscle-specific RING finger proteins (MURFs), in striated muscle. J Muscle Res Cell Motil. 2005;26(6-8):389–400. [PubMed: 16477476]
- 56.
- McElhinny AS, Kakinuma K, Sorimachi H, et al. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J Cell Biol. 2002;157(1):125–136. [PMC free article: PMC2173255] [PubMed: 11927605]
- 57.
- Kedar V, McDonough H, Ary R, et al. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA. 2004;101(52):18135–18140. [PMC free article: PMC539735] [PubMed: 15601779]
- 58.
- Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. [PubMed: 11679633]
- 59.
- Clarke BA, Drujan D, Willis MS, et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metabolism. 2007;6(5):376–385. [PubMed: 17983583]
- 60.
- Stremlau M, Owens CM, Perron MJ, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. [PubMed: 14985764]
- 61.
- Sakuma R, Noser JA, Ohmine S, et al. Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat Med. 2007;13(5):631–635. [PubMed: 17435772]
- 62.
- Hatziioannou T, Perez-Caballero D, Yang A, et al. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5. Proc Natl Acad Sci USA. 2004;101(29):10774–10779. [PMC free article: PMC490010] [PubMed: 15249685]
- 63.
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5a genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci USA. 2004;101(29):10780–10785. [PMC free article: PMC490011] [PubMed: 15249687]
- 64.
- Perez-Caballero D, Hatziioannou T, Yang A, et al. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79(14):8969–8978. [PMC free article: PMC1168745] [PubMed: 15994791]
- 65.
- Sebastian S, Luban J. Retrovirology. Vol. 2. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid; p. 40. [PMC free article: PMC1166576] [PubMed: 15967037]
- 66.
- Diaz-Griffero F, Li X, Javanbakht H, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349(2):300–315. [PubMed: 16472833]
- 67.
- Asaoka K, Ikeda K, Hishinuma T, et al. A retrovirus restriction factor TRIM5a is transcriptionally regulated by interferons. Biochem Biophys Res Commun. 2005;338(4):1950–1956. [PubMed: 16289103]
Publication Details
Author Information and Affiliations
Authors
Kazuhiro Ikeda1 and Satoshi Inoue1,2,*.Affiliations
Notes
Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Ikeda K, Inoue S. TRIM PROTEINS AS RING FINGER E3 UBIQUITIN LIGASES. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.