?
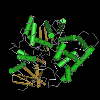 
cytochrome P450 family 107 and similar cytochrome P450s This group contains bacterial cytochrome P450s from families 107 (CYP107), 154 (CYP154), 197 (CYP197), and similar proteins. Among the members of this group are: Pseudonocardia autotrophica vitamin D(3) 25-hydroxylase (also known as CYP197A; EC 1.14.15.15) that catalyzes the hydroxylation of vitamin D(3) into 25-hydroxyvitamin D(3) and 1-alpha,25-dihydroxyvitamin D(3), its physiologically active forms; Saccharopolyspora erythraea CYP107A1, also called P450eryF or 6-deoxyerythronolide B hydroxylase (EC 1.14.15.35), that catalyzes the conversion of 6-deoxyerythronolide B (6-DEB) to erythronolide B (EB) by the insertion of an oxygen at the 6S position of 6-DEB; Bacillus megaterium CYP107DY1 that displays C6-hydroxylation activity towards mevastatin to produce pravastatin; Streptomyces coelicolor CYP154C1 that shows activity towards 12- and 14-membered ring macrolactones in vitro and may be involved in catalyzing the site-specific oxidation of the precursors to macrolide antibiotics, which introduces regiochemical diversity into the macrolide ring system; and Nocardia farcinica CYP154C5 that acts on steroids with regioselectivity and stereoselectivity, converting various pregnans and androstans to yield 16 alpha-hydroxylated steroid products. Bacillus subtilis CYP107H1 is not included in this group. The CYP107-like group belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
