Resequencing
Introduction
Resequencing of candidate genes or other genomic regions of interest in patients and controls is a key step in detection of mutations associated with various congenital diseases. Resequencing techniques can be divided into those which test for known mutations (genotyping) and those which scan for any mutation in a given target region (variation analysis). Typical mutations being tested are substitution (SNP), insertion and deletion mutations.
Eectrophoresis-based resequencing.
Electrophoresis is a DNA separation technique that is very important in DNA sequencing. Standard sequencing procedures involve cloning DNA fragments into special sequencing cloning vectors that carry tiny pieces of DNA. The next step is to determine the base sequence of the fragments by generating a series of even tinier DNA fragments that differ in size by only one base. These fragments are separated by gel electrophoresis, in which the DNA pieces work their way down through the polymer matrix which is placed under electrical field. Smaller pieces move faster and will reach the bottom first.
One of most advanced resequencing techniques based on electrophoresis is capillary electrophoresis. VariantSEQr™ system created by Applied Biosystems successfully integrates capillary electrophoresis, PCR and state-of-the-art automated data analysis techniques for quick and accurate resequencing of specific human genes.
Array-based high-throughput resequencing strategies.

I. The gain of hybridization signal approach.
In the gain of hybridization signal approach, relative hybridization to allele-specific probes complementary to each of the four possible nucleotides at interrogated nucleotide position is used for genotype analysis.
Disadvantage: analysis requires large amount of carefully designed probes, for example, interrogating both target strands of length N for all possible insertions of length X requires 2(4X)N probes.
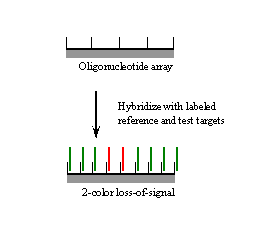
II. The loss of hybridization signal approach.
In the loss of hybridization signal approach, decreased hybridization of red-labeled test target relative to green-labeled reference target to perfect match probes interrogating the area of interest indicates the presence of a sequence change.
Disadvantage: the mutation cannot be discerned; the identity of the sequence change must be established by subsequent dideoxysequencing of the region surrounding the loss of signal signature.
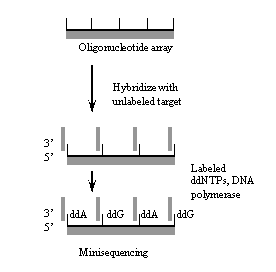
III. The minisequencing approach.
In the minisequencing approach unlabeled target is hybridized to perfect match probes (attached through a 5' linkage to the array to leave an exposed 3'-OH group) interrogating the nucleotide position of interest. Fluorescently tagged ddNTPs are used in subsequent enzymatic primer extension reactions to extend the hybridized primers. The identity of the extended ddNTP is used in sequence analysis. Disadvantage: designing and validating primers can be a long, tedious process that often leads to experimental delays and defective PCR products; data analysis requires not only a high level of expertise but also substantial time commitment.
Sample Queries
Resources
» VariantSEQr™ Resequencing Project
» About resequencing in PubMed

|
|

|
Disclaimer
Mention of specific products or vendors on this website does not constitute an endorsement by the U.S. government.
