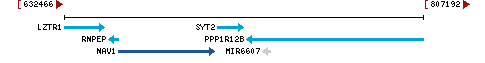- Exon count:
- 34
| Annotation release |
Status |
Assembly |
Chr
|
Location |
|
106
|
current |
bGalGal1.mat.broiler.GRCg7b (GCF_016699485.2) |
26 |
NC_052557.1 (658930..705344) |
|
106
|
current |
bGalGal1.pat.whiteleghornlayer.GRCg7w (GCF_016700215.2) |
26 |
NC_052598.1 (628182..674693) |
Chromosome 26 - NC_052557.1