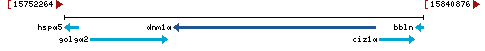- Location:
-
chromosome: LG14
- Exon count:
- 25
| Annotation release |
Status |
Assembly |
LG
|
Location |
|
102
|
current |
ASM164957v2 (GCF_001649575.2) |
LG14 |
NC_051443.1 (15779152..15829045, complement) |
|
101
|
previous assembly |
ASM164957v1 (GCF_001649575.1) |
Unplaced Scaffold |
NW_016094249.1 (5428831..5478710, complement) |
Linkage Group LG14 - NC_051443.1