- Exon count:
- 35
| Annotation release |
Status |
Assembly |
Chr
|
Location |
|
106
|
current |
Sscrofa11.1 (GCF_000003025.6) |
9 |
NC_010451.4 (7136637..7227966, complement) |
| 105 |
previous assembly |
Sscrofa10.2 (GCF_000003025.5) |
9 |
NC_010451.3 (7896906..7988379, complement) |
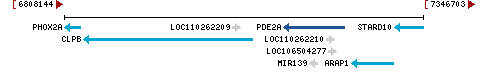
Chromosome 9 - NC_010451.4