CtBP family proteins are unique in animals and in plants. The invertebrates and plants contain a single CtBP family gene while vertebrates have two genes. Genetic studies in Drosophila and in mice indicate that CtBPs play pivotal roles in animal development. The vertebrate CtBPs (CtBP1 and CtBP2) are highly related and are functionally redundant for certain developmental processes and non redundant for others. The vertebrates code two isoforms of each CtBP1 and CtBP2. The animal CtBPs exhibit a highly conserved sequence and structural similarity to D-isomer specific 2-hydroxy acid dehydrogenases (D2-HDH). Structural and molecular modeling studies indicate that CtBP1 is a dehydrogenase and could also function as a lysophosphatidic acyl-CoA transferase under a different configuration. The CtBP family members function predominantly as transcriptional corepressors in the nucleus in conjunction with a number of different DNA binding repressors. The transcriptional regulatory activity of CtBPs appears to be regulated by NAD(H)-binding and the metabolic status of the cell. The corepressor complex of CtBP1 contains enzymatic constituents that mediate coordinated histone modification by deacetylation and methylation of histone H3-K9 and demethylation of histone H3-K4. In the cytosol, they perform diverse functions associated with membrane trafficking, central nervous system synapses and in regulation of the microtubule cytoskeleton. The mammalian CtBPs modulate oncogenesis by regulating the activities of tumor suppressor genes and cellular and viral oncogenes, consistent with a role in tumor suppression as well as in tumor promotion. The CtBPs promote tumorigenesis by repressing transcription of several critical pro-apoptotic genes and by inhibiting genes involved in the epithelial to mesenchymal transition. This Chapter presents a comprehensive general review of the CtBP field and highlights contents of the individual Chapters of this book which contain detailed discussions on structure and functions of animal and plant CtBP family proteins.
Introduction
CtBP (C-terminal binding protein) was identified in 1993 as a 48 kD cellular phosphoprotein that bound to the C-terminal region of the adenovirus E1A oncoprotein.1 In 1995, the cDNA for the founding member of the CtBP family protein was cloned and the encoded protein was shown to bind to a five amino acid motif (PLDLS) conserved at the C-terminus of E1A of all primate adenoviruses.2 The CtBP protein originally identified as the E1A-binding protein is now known as CtBP1. Subsequently, a highly homologous human protein termed CtBP2 was identified by analysis of EST data bank sequences3 and mouse CtBP2 was cloned by a two hybrid screen against the transcription factor BKLF.4 The initial amino acid homology searches revealed that CtBP1 shared a striking homology to D-isomer specific 2-hydroxy acid dehydrogenases (D2-HDH).2 The interaction between a cellular protein with a metabolic enzyme fold and the E1A viral oncoprotein was unexpected since E1A functions primarily as a transcriptional modulator (reviewed by Gallimore and Turnell).5 However, a possible role of CtBP in transcriptional repression was soon suggested by a tethering transcriptional assay.6 In these assays, the N-terminal conserved region (CR1) of E1A fused to a heterologous DNA-binding domain (Gal4) strongly activated a synthetic promoter containing a Gal4 binding site. The CR1 region of E1A contains the sequences for interaction with a SWI/SNF-related chromatin remodeling complex, TRRAP/p4007-9 and also the binding sites for the nuclear acetylase P/CAF.10 Inclusion of the C-terminal region of E1A in the chimeric Gal4-E1A construct abrogated CR1-mediated transcriptional activation. Deletion of the CtBP-binding motif relieved the repressive activity of the C-terminal region. These results suggested that interaction of CtBP with the C-terminal region antagonized the trans-activation activity of CR1 in cis.
A definitive role for CtBP in transcriptional repression became evident with the identification and cloning of the Drosophila homolog of CtBP (dCtBP) by the laboratories of Michael Levine11 and Susan Parkhurst.12 Since then, a large number of DNA-binding transcriptional repressors have been reported to recruit CtBPs via the PLDLS-related binding sites.13,14 The studies with dCtBP and a number of subsequent studies with vertebrate CtBP1 and CtBP2 have established that CtBPs function predominantly as transcriptional corepressors. However, splice variants of the vertebrate CtBPs have been shown to be involved in unrelated biological processes in the cytosol. During the past ten years since the cloning of CtBP1, there has been a substantial increase in our understanding of the structure, functions, and mechanisms of action of CtBP family proteins and their role in various biological processes. These advancements include elucidation of the structural determinants of CtBP1 and the molecular basis of its interaction with the CtBP-binding motif and the determination of the roles of CtBP1 and CtBP2 in mouse development. Additionally, several nuclear cofactors that mediate the transcriptional regulatory activity of CtBPs and the CtBP-target genes have been identified. This Chapter will highlight the salient aspects of CtBP family proteins while more detailed discussions can be found in the individual Chapters of this book.
CtBP Family Proteins
The CtBP family proteins are highly conserved in higher eukaryotes. The genomes of invertebrates such as Drosophila and C. elegans contain a single CtBP gene. However, they code different isoforms as a result of differential RNA processing. For example, in Drosophila there appears to be at least three different alternatively spliced transcripts of dCtBP15 (see Chapter 2). The vertebrate genomes contain two different genes, CtBP1 and CtBP2 that code for two highly related proteins. The CtBP1 gene is located on chromosome 4 of humans and on chromosome 5 of mice. In mammals, the CtBP1 gene expresses two major transcripts as a result of alternate RNA splicing. These transcripts encode two isoforms of CtBP1, which are identical except for a thirteen amino acid region at the N-terminus (Fig. 1A). The short version of CtBP1 (CtBP1-S) corresponds to an isoform designated as CtBP3/BARS16 (see Chapter 10; the designation CtBP3 has now been changed to CtBP1-S). The transcript for CtBP1-S has an alternate inframe exon (exon 2) in the 5'-region which codes for the N-terminal two amino acids while translation of CtBP1-L is initiated from exon 1 of the shorter transcript (Fig. 1A). A fraction of CtBP1 cDNAs also contains an insertion of a codon for a Ser residue (at position 380 in CtBP1-L and at position 369 in CtBP1-S), which also appears to be the result of alternate RNA processing. The functional significance of the extra Ser residue is not known at present. The CtBP1 proteins are concentrated in the nucleus with significant amounts in the cytosol.
The CtBP2 gene maps to chromosome 10 of humans and in chromosome 7 of mice. The CtBP2 gene also codes for two protein isoforms (Fig. 1B). The ubiquitously expressed isoform commonly referred to as CtBP2 (48 kD) is highly related to CtBP1. The second isoform, designated as RIBEYE is a 120 kD protein and is predominantly expressed in sensory neurons.17,18 The two protein isoforms of the CtBP2 gene are coded by two transcripts that are transcribed from two distinct promoters and contain two different 5'-coding exons.17 The first coding exon of the CtBP2 transcript codes for the N-terminal 20 amino acids of CtBP2 while the first coding exon of the RIBEYE transcript (located within the first intronic region of the CtBP2 locus) codes for a large N-terminal domain (designated A-domain) of RIBEYE. The A-domain of RIBEYE is unrelated to other proteins and the B-domain is identical to CtBP2 (aa 21 to 445). RIBEYE lacks the N-terminal 20 amino acid domain of CtBP2 and is localized to the cytoplasm.19 In contrast, CtBP2 is highly concentrated in the nucleus (L. Zhao and G.C., unpublished). The CtBP2 gene is unique since its transcription is controlled by two distinct promoters to generate transcripts for two different protein isoforms. In contrast to mammals, the telocast fish express two different RIBEYE proteins coded by two different CtBP2 genes.20 The RIBEYE protein, in concert with other protein factors which includes CtBP1, plays a central role in ribbon synapses.
The genomes of terrestrial plants also code for a CtBP family member, ANGUSTIFOLIA (AN). The AN gene was first identified in Arabidopsis thaliana21,22 and this gene controls polarity-dependent leaf cell expansion, possibly through controlling the arrangement of the microtubule cytoskeleton (see Chapter 12 by H. Tsukaya). The N-terminal half of the AN protein shares amino acid sequence homology with animal CtBP family members. Although the sequence conservation between AN and animal CtBPs is not as extensive as among the animal CtBPs, the shared homology between AN and CtBPs is significant and warrants inclusion of AN in the CtBP family. However, some differences between AN and animal CtBPs are evident. In spite of a general sequence similarity to D2-HDH, the AN protein lacks amino acid residues important for the D2-HDH catalytic function and also lacks the consensus NAD(H)-binding motif. In contrast to animal CtBPs, the AN protein of A. thaliana does not appear to bind to the prototypical PLDLS motif containing protein E1A (see Chapter 12). Plant AN proteins contain a conserved Rb-binding motif at the N-terminal region. However, it is not known if AN complexes with Rb family members. Like animal CtBPs, AN also appears to be both cytoplasmic and nuclear suggesting that it functions at both locations. Genetic and biochemical studies have indicated an association between the AN protein and the kinesin motor ZWICHEL (ZWI) consistent with a role for AN in control of the microtubule cytoskeleton.21 Gene expression profiling studies suggest that AN might also function as a transcriptional corepressor since expression of a set of genes was elevated in an mutant plants.22 Since some functionally deficient an mutations are located in the C-terminal unique region of the AN gene (not present in animal CtBPs), it appears that AN may have functions in addition to those that are controlled by the CtBP-homology region. Results from future studies are eagerly awaited to determine if the CtBP-homology region of AN can be substituted by animal CtBP sequences.
Nuclear Functions
Transcriptional Repression
The vertebrate CtBPs4,6,23-25 and the Drosophila homolog, dCtBP11,12 function as transcriptional corepressors in the nucleus. It is now well established that a large number of DNA-binding transcriptional repressors mediate their activity by recruiting CtBP through sequence motifs that resemble the adenovirus E1A CtBP-binding motif, PLDLS.13,14 Initial studies with Drosophila embryos provided strong evidence that dCtBP is a transcriptional corepressor.11 Since then a number of Drosophila repressors have been shown to mediate their activity partly or fully in a dCtBP-dependent manner (see Chapter 2 by Aihara, Perrone and Nibu). These conclusions were based on two different approaches. First, in embryos deficient in maternal dCtBP, the activities of several repressors were impaired. Second, in the transgenic embryos, repressors mutated in the CtBP-binding motif were defective in transcriptional repression. Although dCtBP interacts with a long range repressor, Hairy,12 it appears that dCtBP may inhibit the repressive function of Hairy by modulating the activity of the corepressor Groucho (Gro) (which contributes to the repressor activity of Hairy).26 The manifestation of full activity of short range repressors such as Krüppel Knirps, Snail and Giant requires dCtBP.11,27 CtBP appears to contribute quantitatively, rather than qualitatively, to the activity of short range repressors such as Knirps.28 The molecular mechanism by which dCtBP contributes transcriptional repression in conjunction with these repressors remains to be elucidated. Although hCtBP1 associates with type I histone deacetylases (HDAC),29-31 it is uncertain whether dCtBP associates with HDAC since short range repressors function normally in mutant embryos that are deficient in dHDAC132 and are insensitive to trichostatin A.33 However, a more recent analysis of a protein complex of the short-range repressed Knirps has revealed the presence of Rpd3 and this association requires the CtBP-dependent repression domain of Knirps.33a Thus, the possibility that dCtBP-mediated short-range repression requires the HDAC activity warrant further scrutiny.
Mutational studies indicate that the putative D2-HDH activity of dCtBP is not required for transcriptional repression while the NAD(H)-binding activity is required when expressed as the DNA-binding Gal4-dCtBP fusion protein.15 In addition to the role in short range repression, recent studies with transgenic Drosophila embryos have provided strong evidence that dCtBP plays a critical role in repression mediated by the Polycomb group (PcG) proteins.34,35 The mammalian transcriptional repressor YY1 which shares significant sequence homology to the Drosophila PcG protein Pleiohomeotic (PHO) represses PcG-responsive promoters when expressed as a Gal4-YY1 chimeric protein. The YY1-mediated repression is strongly dependent on dCtBP. These studies have revealed that dCtBP plays direct role in PcG repression by modulating the DNA-binding activity of Gal4-YY1 as well as by recruiting other PcG factors.
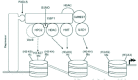
With regard to the subcellular localization of the two vertebrate CtBP proteins, CtBP1 localizes to both the nucleus and the cytoplasm, with enhanced nuclear concentration. In contrast, CtBP2 localizes predominantly to the nucleus. Consistent with the high degree of sequence homology shared by these two proteins, both proteins have been shown to possess transcriptional corepressor activity. The sequence conservation would also suggest that they might mediate the transcriptional repression activity through similar mechanisms. Studies with mutant mice also suggest that the two isoforms have overlapping transcriptional functions.36 Most of our current knowledge on transcriptional regulation by vertebrate CtBPs is derived from studies using CtBP1 as the model. Although structural studies have established that CtBP1 is a D2-HDH37,38 and biochemical studies indicate that CtBP1 possess a slow DH activity29,37,39 (see Chapter 9 by Lundblad), the role of DH activity in transcriptional repression by vertebrate CtBPs is not clear and remains controversial.4,37,40 A proteomics based analysis of the CtBP nuclear protein complex by Yang Shi and colleagues has illuminated some critical aspects of the corepressor function of CtBP129 (see Chapter 8 by Shi and Shi). These studies have identified several chromatin modifying enzymatic constituents associated with the CtBP protein complex, in addition to certain DNA-binding repressors (such as ZEB) that have been previously known to recruit CtBP. The CtBP complex contains class 1 histone deacetylases (HDAC1/2) and histone methylases (G9a and HMTase1) suggesting that CtBP1 contributes to transcriptional repression by coordinate histone modification through deacetylation and methylation (Fig. 2). Studies on the CtBP protein complex have also led to the identification of the first histone demethylase, LSD1 (lysine specific demethylase-1). Additionally, the CtBP complex contains the corepressor CoREST.41,42 The CoREST protein complex also contains HDAC1/2 and LSD1 (BHC110).43 It appears that CoREST is the direct binding partner of LSD1.44,45 In Chapter 8, Shi and Shi suggest that the CoREST repressor complex may be substantially similar to the CtBP complex since they share a number of constituents (such as HDAC1/2 and LSD1), and repress a common set of target genes.
The CtBP protein complex also contains the PcG protein HPC2. The interaction between PC2 and CtBP has also been detected previously in two hybrid screenings.25 The HPC2 protein recruits CtBP1 and Ubc9 to the PcG bodies46 resulting in sumoylation of CtBP1 at a single Lys (K428) residue.46,47 The SUMO modification of CtBP1 appears to be critical for its nuclear accumulation. The potential role of HPC2 in CtBP-mediated transcriptional repression remains to be investigated. In Chapter 8, the authors raise the possibility that HPC2 may function by binding to methylated (K27) histone H3 in a fashion analogous to the Drosophila Pc protein. Although sumoylation of CtBP1 has been reported to be important for nuclear localization, it is possible that this modification may also play a role in the corepressor activity of CtBP1. Certain transcription factors have been reported to recruit HDACs via the SUMO peptide.48,49 Additionally, Ubc9 recruited by CtBP1 and HPC2 may also target other transcription factors and histone H4.50-52 Since the SUMO peptide has been shown to bind HDACs such as HADC2 and 6 (reviewed by Gill),53 it would be of interest to determine if the recruitment of HDACs by CtBP1 is dependent on sumoylation. It is also possible that HDAC1/ 2 may be recruited to the CtBP complex through CoREST. Thus, studies on the CtBP protein complex suggest that coordinate histone modification may be the primary mode of transcriptional repression by CtBP1. However, CtBP1 has also been reported to inhibit the general transcriptional machinery through direct interaction with nuclear acetylases p300 and CBP via a PXDLS motif located within the bromodomain of these enzymes,54a as well as through a PXDLS-independent interaction.55
As in the case of dCtBP, the dinucleotide binding activity of vertebrate CtBPs also plays an important role in transcriptional activity. The dinucleotides NAD+ and NADH stimulate dimerization and interaction of CtBP with PXDLS-containing target proteins such as adenovirus E1A.39,56 Since CtBP1 appears to show enhanced affinity for NADH than to NAD+, CtBP has been postulated to be a redox sensor that links the cellular metabolic status to transcriptional regulation.57 In Chapter 7, Goodman, Zhang and colleagues discuss the experimental and structural evidences in support of this model. Interestingly, it is possible that there may be a functional relationship between CtBP and the dinucleotide-dependent transcriptional regulator Sir2. The potential relationship between these two transcriptional coregulators has been suggested by a chromatin profiling study in Drosophila embryos where 90% of dSir2- recruiting loci were also found to recruit dCtBP.57a
Transcriptional Activation
Although CtBPs function predominantly as transcriptional corepressors linking various chromatin-modifying components to DNA-binding repressors, under certain conditions they may function as transcriptional activators. Studies with CtBP2-null mouse embryos which exhibit axial truncation phenotypes have revealed that expression of one of the target genes of Wnt3A, Brachyury, is lower in E10.5 embryos compared to normal looking E9.5 embryos.36 This observation suggests that CtBP2 may function as a transcriptional activator of Brachyury. A context-specific transcriptional activation function for dCtBP has also been suggested based on transcriptional tethering studies with Gal4-dCtBP in different mammalian cell lines.58 In contrast to the repression function, it appears that the activation function of CtBP may be indirect. For example, mTcf3, which represses Brachyury primarily through the corepressor Gro, contains two divergent CtBP-binding motifs. It is possible that CtBP may activate Brachyury by interfering with the repressive function of Gro. It should be noted that a similar antagonism between Gro and CtBP has been observed in the context of transcriptional regulation by Hairy in Drosophila.59
Cytosolic Functions of CtBP
Role in Membrane Fission and Transport
CtBP1 has also been identified as a 50 kD cytosolic target (designated BARS-50) for ribosylation that is mediated by the fungal toxin brefeldin A (BFA) in the Golgi.60 The ability of BFA to disassemble Golgi appears to correlate with ribosylation of BARS-50. Protein purification and cDNA cloning identified the rat homolog of CtBP1 as BARS-5016 (see Chapter 10 by Spano, Hidalgo Carcedo, and Corda). Although the cloned cDNA corresponds to the splice variant CtBP1-S (Fig. 1), it appears that both isoforms of CtBP1 (CtBP1-L and CtBP1-S) may have BARS activity. In vitro studies have revealed that recombinant or purified CtBP/BARS can induce fission of isolated Golgi membrane.61 During these studies, CtBP1-S was shown to bind acyl-CoA and possess a slow acyltransferase (LPAAT) activity, which selectively catalyzes acylation of lysophosphatidic acid (LPA) to phosphatidic acid (PA) using acyl-CoA.61 In Chapter 10, Spano et al discuss the structural basis for the dual function of CtBP1 (also see Chapter 9 by Lundblad). Based on molecular modeling,38 they suggest that CtBP1 exhibits fissioning activity when it binds acyl-CoA and assumes an open structural configuration as a monomer and participates in transcriptional regulation when in the dimeric form bound to NAD(H). Although the initial studies ascribed the Golgi membrane fission activity of CtBP/BARS to the acyltransferase activity,61 subsequent studies have suggested that this activity plays only an enhancing role since a mutant of CtBP/BARS defective in the enzyme activity was able to induce membrane fission with lower efficiency.62 Studies using mitotic cytosolic extracts from normal rat kidney (NRK) cells that were immuno-depleted for CtBP/BARS and then reconstituted with recombinant wt or dominant negative mutants of CtBP/BARS revealed that CtBP is important for the mitotic fragmentation of the Golgi complex.62 These results have been further extended using living cells that were microinjected with CtBP antibodies or dominant negative mutants or antisense oligonucleotides. The results from such studies have suggested a critical role for CtBP1 in mitotic partitioning of Golgi in the NRK model.
Since fission is a critical step in membrane transport, Luini, Corda and colleagues have also investigated the role of CtBP/BARS in the formation of transport carriers from the Golgi complex to the plasma membrane.63 Both siRNA-mediated depletion of CtBP1 and the same approaches used to demonstrate a role in mitotic partitioning of Golgi complex were used to demonstrate a role for CtBP1 in dynamin-independent endocytic and exocytic transport pathways in cells of epithelial origin. In contrast to the results on membrane fission and transport obtained with NRK and COS (monkey kidney) cells, studies with mouse embryo fibroblasts (MEF) that are null for both CtBP1 and CtBP236 do not appear to show any significant defects in Golgi partitioning. These MEFs also appear to proliferate normally and are not deficient in membrane transport (see Chapter 6 by Hildebrand).63 Similarly, the AN mutants of A. thaliana also do not appear to show any Golgi defects (see Chapter 12). In Chapter 10, Spano et al discuss the possibility that ‘adaptive’ mechanisms during embryonic development might have caused the CtBP-independent Golgi partitioning and transport mechanisms observed in CtBP-null MEF. The cell types could also be a critical determinant for the requirement of CtBP1 for Golgi fission and transport. An interesting question is whether CtBP2 could substitute for CtBP1 in the membrane fission and transport assays. The membrane fission and transport activities mediated by CtBPs may have relevance to central nervous system synapses (see below). This issue is addressed by tom Dieck et al in Chapter 11.
CtBPs in Central Nervous System Synapses
The discovery and cloning of RIBEYE (Fig. 1B) as a component of the ribbon synaptic complex revealed a surprising function for CtBPs in central nervous system synapses.17 Visual and auditory sensory neurons are endowed with the capacity for tonic release of the neurotransmitter. These cells express a synaptic ‘ribbon’ that tethers clusters of vesicles and transports them to active sites at the plasma membrane (Fig. 3). Although several proteins have been identified in the ribbon complex, RIBEYE appears to be a major constituent.17 While the B-domain (CtBP2) of RIBEYE is highly conserved between species, the A-domain is divergent, suggesting that the A-domain plays a ribbon-specific structural role in forming the ribbon backbone. Depletion of RIBEYE in zebrafish (by the use of morpholino antisense oligonucleotides) has been shown to result in shorter synaptic ribbons.20 As discussed in Chapter 11, Brandstätter and colleagues have discovered that CtBP1 is also a constituent of the ribbon synapses.19 The role of CtBPs in tethering of vesicles to the ribbon and their mobilization appear to be independent of PXDLS binding.18 Each ribbon appears to contain ˜ 4000 molecules (RIBEYE/CtBPs) that bind to a PXDLS-containing fluorescent peptide probe, thus comprising the majority of the volume (›60%) of the ribbon.18 Ultra structural studies have revealed that both RIBEYE and CtBP1 colocalize throughout the ribbon structure. The presence of both RIBEYE and CtBP1 may meet the needs of tonic rate release of neurotransmitter. Brandstätter and colleagues have also identified CtBP1 as a constituent of the conventional chemical synapses that do not express RIBEYE.19 In Chapter 11 tom Dieck et al propose two roles for CtBPs in chemical synapses, a structural role (backbone of ribbon and ribbon variations) and a role in membrane turnover. They suggest that the LPAAT activity that modulates the curvature of lipid membranes may be important for exocytosis of membrane vesicles. The slow LPAAT activity of CtBP1 implicated in Golgi fission61 may be relevant in the mobilization of synaptic vesicles. It would be interesting to know if RIBEYE possesses any LPAAT activity or facilitates recruitment of CtBP1 (via heterodimerization) to the ribbon synapses. The availability of a knockout mouse model for CtBP1 makes it possible to investigate the role of CtBP1 in central nervous system synapses.
Control of Plant Microtubule Cytoskeleton
As discussed in Chapter 12, the AN gene of Arabidopsis thaliana controls the process of leaf hair (trichome) branching and polarized leaf cell expansion, which influences the leaf shape. An abnormal distribution of microtubules is present in AN mutant plants. Genetic studies revealed that the AN gene might interact with the gene ZWICHEL (ZWI),21 which codes for a protein related to the kinesin motor molecule.64 The genetic interaction between AN and ZWI was discovered in double heterozygous plants of certain zwi and an allele combinations. Such heterozygous plants contained more trichome branches than the corresponding wild-types. Further, the zwi mutants exhibited a phenotype similar to the an mutants.65 Yeast two hybrid analysis also indicated a physical interaction between AN and ZWI proteins. It is interesting to note that the mammalian ribbon synaptic complexes also contain a kinesin related protein KIF3A, in addition to CtBPs.19 It has been suggested that some AN functions in leaf cell morphogenesis may be linked to directional vesicle trafficking controlled by the microtubule cytoskeleton and motor molecules such as ZWI.65
Role of CtBPs in Developmental Processes
CtBP family proteins play critical roles during development of both invertebrates and vertebrates. Homozygous inactivation of the dCtBP gene in Drosophila is lethal.12 Embryos with reduced levels of maternal dCtBP exhibit severe segmentation defects,12,27 which have been attributed to the loss of repression of target genes by several short range transcriptional repressors. The Chapter by Aihara, Perrone and Nibu (Chapter 2) discusses the activities of short range repressors and dCtBP in the early Drosophila embryo. Studies with Xenopus embryos have also revealed that CtBPs play critical roles in development by regulating the activities of transcriptional regulators such as Tcf-3, FOG and ZEB-2/SIP1.24, 66 - 68 In Chapter 3, Verger, Perdomo and Crossley discuss the role of FOG and CtBP in hematopoiesis in Xenopus and in Drosophila.
In Chapter 6, Jeffrey Hildebrand discusses his genetic analysis of mice with mutations in the CtBP1 and CtBP2 genes.36 His studies have revealed that the two CtBP isoforms have nonredundant as well as redundant functions during mouse development. CtBP1-null mice are viable but are small and less robust. Homozygous inactivation of the CtBP2 locus results in embryonic lethality between E9 and E10.5, primarily due to defects in placental development. Some of the phenotypes associated with CtBP2-null embryos may be attributed to a reduction in expression of the T-box transcription factor Brachyury. A prominent phenotype associated with the deficiency of CtBP isoforms appears to be the presence of extensive epithelial components in various tissues and organs. This is consistent with the role of CtBP in repressing the expression of various genes important for conferring epithelial phenotype such as E-cadherin.40 Although the pathways controlled by CtBPs during development remain to be clarified in detail, Hildebrand highlights the link between CtBPs and signaling pathways such as the Wnt and TGF-β/BMP during development. A study with Drosophila embryos has revealed a link between dCtBP and modulation of the Wg pathway during development.69 A more recent analysis of the expression of CtBP1 and CtBP2 genes in avian embryonic development also suggest that the two genes may play functionally redundant roles in development of some tissues and unique roles in development others,69 like during mouse embryo development. The chapter by Hildebrand also highlights the similarities between the phenotypes observed in CtBP mutant mice and those of the human syndrome Holoprosencephaly (HPE). Valuable CtBP mutant mouse models should facilitate further elucidation of the roles of CtBPs in vertebrate development.
Role in Oncogenesis and Apoptosis
The available evidence suggests that CtBPs may play important roles in tumorigenesis and tumor progression by modulating the activities of oncogenes, signaling pathways, and apoptosis. A role of CtBP in oncogenesis was first inferred from studies with the adenovirus E1A oncogene.1,2,70 Mutations in the C-terminal region of the E1A protein that obliterate the CtBP-binding motif (PLDLS) induced enhanced transformation of primary rodent epithelial cells in cooperation with the activated Ras oncogene (Fig. 4). Transformed cells expressing the mutant E1A and the Ras oncogene were also highly tumorigenic and metastatic. Thus, the interaction of CtBP with the C-terminus of E1A results in suppression of the full oncogenic activity of the Ras oncogene. It appears that the hyper-transforming phenotype of E1A C-terminal mutants is specific for cooperative transformation with the Ras oncogene, since such E1A mutations are defective in transformation in cooperation with the adenovirus E1B region.71-73 More recent studies by Grand and coworkers (described in Chapter 5), also suggest that a mutation within the CtBP-binding motif confers a temperature sensitive phenotype to Ad12 E1A-EIB cooperative transformation. The role of the C-terminal region of E1A in E1A-E1B cooperative transformation may be linked to the inability of E1A C-terminal mutants to induce immortalization of primary cells. The Ras oncogene may override an immortalization restriction to induce oncogenic transformation in cooperation with E1A C-terminal mutants. The CtBP-binding motif of E1A is implicated in relief of repression of the telomerase (hTERT) promoter.74 It is possible that the immortalization defect of C-terminal (exon 2) mutants of E1A may be linked to their inability to activate the hTERT promoter.
Although the mechanism by which CtBP interaction with E1A modulates oncogenic transformation is not fully understood, it appears that most of this activity may be related to relief of CtBP-mediated transcriptional repression by the second exon of E1A. Frisch and coworkers have demonstrated that E1A induced expression of several epithelial genes and E1A mutants defective in interaction with CtBP were partially deficient in activation of these genes.75 A microarray analysis, which compared the gene expression profiles of cells expressing wt E1A or an E1A mutant lacking the CtBP-binding motif, identified a number of genes that were activated by wt E1A and not by the C-terminal mutant.76 These genes included those involved in tumor progression and growth suppression.76 A different gene expression profiling study using CtBP-null mouse embryo fibroblasts (MEF) and CtBP1-rescued MEF has revealed that several epithelial (such as cytokeratins, tight junction components and lamins) and pro-apoptotic (such as PERP, Noxa and Bax) genes are activated in the absence of CtBP40 (see Chapter 4 by Frisch). Thus, it appears that the enhanced transforming properties of E1A mutants may be related their inability to relieve CtBP-mediated repression. The activated Ras oncogene is known to induce epithelial to mesenchymal transition (EMT) with loss of membranous E-cadherin expression.77 The hypertransforming (in cooperation with oncogenic Ras) mutants of E1A may permit unimpeded propagation of Ras activity in modulating EMT.
The above-mentioned studies that suggest the C-terminal region of E1A may serve as a tool to inactivate the transcriptional functions of CtBPs. It is possible that the second exon of E1A that includes the PLDLS motif could be exploited as a therapeutic agent for certain malignancies in which the expression of tumor-restraining genes is repressed by CtBP-dependent repressors. A cellular protein, Pinin/DRS (Pnn), implicated in mRNA processing has also been reported to relieve CtBP-mediated repression of E-cadherin.78 It is possible that Pnn may modulate oncogenesis by regulating EMT in a fashion analogous to E1A. It appears that certain apoptotic stimuli may also mimic the effect of proteins such as E1A and Pnn in neutralizing CtBP functions. Goodman, Zhang and coworkers have shown that in response to exposure to UV, the homeodomain-interacting protein kinase (HIPK2) phosphorylates CtBP1 at Ser-422 resulting in rapid ubiquitination and degradation of CtBP1,79,80 which is accompanied by apoptosis. It would be interesting to know if phosphorylation-mediated clearance of CtBP-1 also results in activation of the various pro-apoptotic genes (i.e., PERP, Noxa, and Bax) that are activated in CtBP knockout cells.
Among cellular oncogenes, the activity of Evi-1 is modulated by direct interaction with CtBP. The expression of the Evi-1 oncogene is activated in human myeloid leukemia and myelodysplastic syndromes.81 It is also expressed as a t(3;21) fusion product with AML-1 in chronic myelocytic leukemia.82 Evi-1 inhibits Smad-activated transcription of TGF-β/activin/BMP (bone morphogenetic protein)-responsive genes by recruiting CtBP.83-86 In Chapter 3, Verger, Perdomo, and Crossley discuss the role of CtBP in conjunction with the Evi-1 gene, the AML/Evi-1 fusion gene , the more recently discovered AML1/FOG-2 fusion gene and the MLL (mixed lineage leukemia) gene in leukemogenesis. A direct interaction of CtBP with the viral oncogenes EBNA3A and EBNA3C is also required for the immortalization and cooperative transformation activities of the EBV oncogenes.87,88
Gene expression studies in CtBP-null cells and certain protein interaction studies raise the possibility that there may be some cross-talk between the p53 and CtBP pathways in modulating oncogenesis. The pro-apoptotic genes, PERP, Noxa, and Bax as well as p21 are well known target genes for p53. The expression of these genes was shown to be highly activated in CtBP-null MEFs.40 However, reporter-based assays (using p53-responsive promoter constructs) performed in the MEFs have suggested that CtBPs may not directly antagonize the activity of p53. A different protein interaction study identified interaction between Hdm2 and CtBP2.89 The Hdm2/Mdm2 oncoprotein is known to mediate its oncogenic activity by inactivating p53 through multiple mechanisms and is also known to possess an intrinsic transcriptional repressor activity.90 Based on these results, it has been suggested that CtBP2 may cause promoter-selective inhibition of transcription of p53-responsive genes through interaction with Hdm2/Mdm2.89 The potential link between the p53 pathways and CtBP pathways merit further investigation.
In addition to the potential tumor-promoting activities of CtBP by regulating the activities of oncogenes and tumor suppressor genes, the available evidence also suggests that CtBP has a role in tumor suppression in the colon. During a search for Drosophila proteins that complex with the E-APC (adenomatous polyposis coli) protein, Hamada and Bienz identified dCtBP as an APC-interacting protein.69 They extended these results to the human APC, an important tumor suppressor in the colon. Hamada and Bienz have demonstrated that CtBP binds directly to APC through PXDLS-like motifs conserved between the fly and mammalian APC proteins. The interaction between APC and CtBP results in sequestration of the APC/β-catenin complex, thereby redirecting free nuclear β-catenin away from the Wnt transcription factor hTcf-4 (Fig. 5). Functional cooperation between CtBP and APC is consistent with the model that CtBP may serve a tumor suppressor role in the colon. The observation that some colon cancer cell lines (e.g., COLO320) express APC truncations lacking the CtBP-binding sequences lends support to this view.
Concluding Remarks
In the last ten years since the cloning of the founding member of the CtBP family proteins, these proteins have evolved from an enigmatic state to a state of much biological importance. A number of critical studies, particularly with dCtBP have been instrumental in establishing a clear role for CtBP family members in regulating transcription. Similarly, the characterization of the CtBP super complex from human cells has been a significant advancement towards understanding the mechanisms of transcriptional repression in mammalian cells. Since the mammalian CtBP1 protein complex appears to contain unique constituents not present in the dCtBP complex, future studies will illuminate a common mechanism shared by the invertebrate and vertebrate CtBPs. Future studies are needed to establish a clear transcriptional role for the plant homolog, ANGUSTIFOLIA. Although CtBPs were the first transcriptional regulators identified to also contain a metabolic enzyme fold, recently such enzymatic constituents have been identified in various transcription complexes. Since there is an absolute conservation of the D2-HDH fold among the animal CtBPs, the next challenge would be to identify the relevant substrate(s) for these enzymes. Detailed investigation into the potential regulation by the NAD(H) dinucleotides of the transcriptional activities of CtBP in tumor cells would be important to gain insight into designing strategies for anti-cancer therapeutic intervention and to discover potential cross-talks between pathways controlled by other dinucleotide-regulated transcriptional regulators such as Sir2. The studies on membrane associated functions of CtBP1 have been instrumental in unraveling the dual activity of CtBP1. Compelling evidence that CtBPs also function as synaptic proteins warrants additional investigations on the role of CtBPs in membrane turnover. Apart from the functional importance of the CtBP family proteins, the genomic organization of these genes have illuminated novel strategies employed by vertebrates to encode proteins (e.g., RIBEYE and CtBP2) of diverse functions within a single gene locus to achieve genome compaction. The roles of CtBP in modulating oncogenic outcomes via EMT and apoptosis raise a promising possibility that CtBPs may be good anti-neoplastic drug targets.
Acknowledgements
The author thanks all the contributors to this book. He also thanks Maurice Green, Drew Lichtenstein, Helmut Brandstätter, Yang Shi, Daniela Corda, Steve Frisch, Jim Lundblad and members of Chinnadurai laboratory for their critical comments on this Chapter. The author expresses his gratitude to S. Vijayalingam for construction of the figures. The author received support from a grant by the National Cancer Institute (CA-84941).
References
- 1.
- Boyd JM, Subramanian T, Schaeper U. et al. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12(2):469–478. [PMC free article: PMC413230] [PubMed: 8440238]
- 2.
- Schaeper U, Boyd JM, Verma S. et al. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92(23):10467–10471. [PMC free article: PMC40632] [PubMed: 7479821]
- 3.
- Katsanis N, Fisher EM. A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics. 1998;47(2):294–299. [PubMed: 9479502]
- 4.
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a corepressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17(17):5129–5140. [PMC free article: PMC1170841] [PubMed: 9724649]
- 5.
- Gallimore PH, Turnell AS. Adenovirus E1A: Remodelling the host cell, a life or death experience. Oncogene. 2001;20(54):7824–7835. [PubMed: 11753665]
- 6.
- Sollerbrant K, Chinnadurai G, Svensson C. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 1996;24(13):2578–2584. [PMC free article: PMC145971] [PubMed: 8692699]
- 7.
- Fuchs M, Gerber J, Drapkin R. et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106(3):297–307. [PubMed: 11509179]
- 8.
- McMahon SB, Van Buskirk HA, Dugan KA. et al. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94(3):363–374. [PubMed: 9708738]
- 9.
- Deleu L, Shellard S, Alevizopoulos K. et al. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20(57):8270–8275. [PubMed: 11781841]
- 10.
- Reid JL, Bannister AJ, Zegerman P. et al. E1A directly binds and regulates the P/CAF acetyltransferase. Embo J. 1998;17(15):4469–4477. [PMC free article: PMC1170778] [PubMed: 9687513]
- 11.
- Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280(5360):101–104. [PubMed: 9525852]
- 12.
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17(7):2067–2078. [PMC free article: PMC1170551] [PubMed: 9524128]
- 13.
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9(2):213–224. [PubMed: 11864595]
- 14.
- Turner J, Crossley M. The CtBP family: Enigmatic and enzymatic transcriptional corepressors. Bioessays. 2001;23(8):683–690. [PubMed: 11494316]
- 15.
- Sutrias-Grau M, Arnosti DN. CtBP contributes quantitatively to Knirps repression activity in an NAD binding-dependent manner. Mol Cell Biol. 2004;24(13):5953–5966. [PMC free article: PMC480900] [PubMed: 15199149]
- 16.
- Spano S, Silletta MG, Colanzi A. et al. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J Biol Chem. 1999;274(25):17705–17710. [PubMed: 10364211]
- 17.
- Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: A protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28(3):857–872. [PubMed: 11163272]
- 18.
- Zenisek D, Horst NK, Merrifield C. et al. Visualizing synaptic ribbons in the living cell. J Neurosci. 2004;24(44):9752–9759. [PMC free article: PMC6730242] [PubMed: 15525760]
- 19.
- tom Dieck S, Altrock WD, Kessels MM. et al. Molecular dissection of the photoreceptor ribbon synapse: Physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J Cell Biol. 2005;168(5):825–836. [PMC free article: PMC2171818] [PubMed: 15728193]
- 20.
- Wan L, Almers W, Chen W. Two ribeye genes in teleosts: The role of Ribeye in ribbon formation and bipolar cell development. J Neurosci. 2005;25(4):941–949. [PMC free article: PMC6725632] [PubMed: 15673675]
- 21.
- Folkers U, Kirik V, Schobinger U. et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002;21(6):1280–1288. [PMC free article: PMC125931] [PubMed: 11889034]
- 22.
- Kim GT, Shoda K, Tsuge T. et al. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002;21(6):1267–1279. [PMC free article: PMC125914] [PubMed: 11889033]
- 23.
- Criqui-Filipe P, Ducret C, Maira SM. et al. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18(12):3392–3403. [PMC free article: PMC1171419] [PubMed: 10369679]
- 24.
- Brannon M, Brown JD, Bates R. et al. XCtBP is a XTcf-3 corepressor with roles throughout Xenopus development. Development. 1999;126(14):3159–3170. [PubMed: 10375506]
- 25.
- Sewalt RG, Gunster MJ, van der Vlag J. et al. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19(1):777–787. [PMC free article: PMC83934] [PubMed: 9858600]
- 26.
- Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96(2):535–540. [PMC free article: PMC15171] [PubMed: 9892668]
- 27.
- Nibu Y, Zhang H, Bajor E. et al. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17(23):7009–7020. [PMC free article: PMC1171049] [PubMed: 9843507]
- 28.
- Struffi P, Corado M, Kulkarni M. et al. Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps. Development. 2004;131(10):2419–2429. [PubMed: 15128671]
- 29.
- Shi Y, Sawada J, Sui G. et al. Coordinated histone modifications mediated by a CtBP corepressor complex. Nature. 2003;422(6933):735–738. [PubMed: 12700765]
- 30.
- Subramanian T, Chinnadurai G. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 2003;540(1-3):255–258. [PubMed: 12681518]
- 31.
- Sundqvist A, Sollerbrant K, Svensson C. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 1998;429(2):183–188. [PubMed: 9650586]
- 32.
- Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96(12):6797–6801. [PMC free article: PMC21995] [PubMed: 10359792]
- 33.
- Ryu JR, Arnosti DN. Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities. Nucleic Acids Res. 2003;31(15):4654–4662. [PMC free article: PMC169881] [PubMed: 12888527]
- 33a.
- Struffi P, Arnosti DN. Functional interaction between the Drosophila Knirps short-range transcriptional repressor and Rpd3 histone deacetylase J Biol Chem 2005. (in press) [PMC free article: PMC1802102] [PubMed: 16186109]
- 34.
- Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18(21):2596–2601. [PMC free article: PMC525539] [PubMed: 15520279]
- 35.
- Atchison L, Ghias A, Wilkinson F. et al. Transcription factor YY1 functions as a PcG protein in vivo. Embo J. 2003;22(6):1347–1358. [PMC free article: PMC151054] [PubMed: 12628927]
- 36.
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22(15):5296–5307. [PMC free article: PMC133942] [PubMed: 12101226]
- 37.
- Kumar V, Carlson JE, Ohgi KA. et al. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10(4):857–869. [PubMed: 12419229]
- 38.
- Nardini M, Spano S, Cericola C. et al. CtBP/BARS: A dual-function protein involved in transcription corepression and Golgi membrane fission. EMBO J. 2003;22(12):3122–3130. [PMC free article: PMC162135] [PubMed: 12805226]
- 39.
- Balasubramanian P, Zhao LJ, Chinnadurai G. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 2003;537(1-3):157–160. [PubMed: 12606049]
- 40.
- Grooteclaes M, Deveraux Q, Hildebrand J. et al. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci USA. 2003;100(8):4568–4573. [PMC free article: PMC153596] [PubMed: 12676992]
- 41.
- You A, Tong JK, Grozinger CM. et al. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98(4):1454–1458. [PMC free article: PMC29278] [PubMed: 11171972]
- 42.
- Ballas N, Battaglioli E, Atouf F. et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31(3):353–365. [PubMed: 11516394]
- 43.
- Hakimi MA, Bochar DA, Chenoweth J. et al. A coreBRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99(11):7420–7425. [PMC free article: PMC124246] [PubMed: 12032298]
- 44.
- Shi YJ, Matson C, Lan F. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005 [PubMed: 16140033]
- 45.
- Lee MG, Wynder C, Cooch N. et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005 [PubMed: 16079794]
- 46.
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113(1):127–137. [PubMed: 12679040]
- 47.
- Lin X, Sun B, Liang M. et al. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol Cell. 2003;11(5):1389–1396. [PubMed: 12769861]
- 48.
- Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13(4):611–617. [PubMed: 14992729]
- 49.
- Girdwood D, Bumpass D, Vaughan OA. et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11(4):1043–1054. [PubMed: 12718889]
- 50.
- Ross S, Best JL, Zon LI. et al. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10(4):831–842. [PubMed: 12419227]
- 51.
- Muller S, Berger M, Lehembre F. et al. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275(18):13321–13329. [PubMed: 10788439]
- 52.
- Holmstrom S, Van Antwerp ME, Iniguez-Lluhi JA. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc Natl Acad Sci USA. 2003;100(26):15758–15763. [PMC free article: PMC307641] [PubMed: 14663148]
- 53.
- Gill G. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 2004;18(17):2046–2059. [PubMed: 15342487]
- 54.
- Kim JH, Cho EJ, Kim ST. et al. CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat Struct Mol Biol. 2005;12(5):423–428. [PubMed: 15834423]
- 54a.
- Metoni AR, Lai CH, Yao TP. A Mechanism of COOH-Terminal Binding Protein- Mediated Repression. Mol Cancer Res. 2005;3(10):575–583. [PubMed: 16254191]
- 55.
- Senyuk V, Sinha KK, Nucifora G. Corepressor CtBP1 interacts with and specifically inhibits CBP activity. Arch Biochem Biophys. 2005 [PubMed: 16122695]
- 56.
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295(5561):1895–1897. [PubMed: 11847309]
- 57.
- Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci USA. 2003;100(16):9202–9207. [PMC free article: PMC170896] [PubMed: 12872005]
- 57a.
- Bianchi-Frias D, Orian A, Delrow JJ. et al. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2(7):E178. [PMC free article: PMC449821] [PubMed: 15252443]
- 58.
- Phippen TM, Sweigart AL, Moniwa M. et al. Drosophila C-terminal binding protein functions as a context-dependent transcriptional cofactor and interferes with both mad and groucho transcriptional repression. J Biol Chem. 2000;275(48):37628–37637. [PubMed: 10973955]
- 59.
- Nibu Y, Zhang H, Levine M. Local action of long-range repressors in the Drosophila embryo. Embo J. 2001;20(9):2246–2253. [PMC free article: PMC125437] [PubMed: 11331590]
- 60.
- Di Girolamo M, Silletta MG, De Matteis MA. et al. Evidence that the 50-kDa substrate of brefeldin A-dependent ADP-ribosylation binds GTP and is modulated by the G-protein beta gamma subunit complex. Proc Natl Acad Sci USA. 1995;92(15):7065–7069. [PMC free article: PMC41472] [PubMed: 7624370]
- 61.
- Weigert R, Silletta MG, Spano S. et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402(6760):429–433. [PubMed: 10586885]
- 62.
- Hidalgo Carcedo C, Bonazzi M, Spano S. et al. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305(5680):93–96. [PubMed: 15232108]
- 63.
- Bonazzi M, Spano S, Turacchio G. et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005;7(6):570–580. [PubMed: 15880102]
- 64.
- Song H, Golovkin M, Reddy AS. et al. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA. 1997;94(1):322–327. [PMC free article: PMC19332] [PubMed: 8990207]
- 65.
- Folkers U, Berger J, Hulskamp M. Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124(19):3779–3786. [PubMed: 9367433]
- 66.
- Deconinck AE, Mead PE, Tevosian SG. et al. FOG acts as a repressor of red blood cell development in Xenopus. Development. 2000;127(10):2031–2040. [PubMed: 10769228]
- 67.
- Lerchner W, Latinkic BV, Remacle JE. et al. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: A study using transgenic frog embryos. Development. 2000;127(12):2729–2739. [PubMed: 10821770]
- 68.
- Postigo AA, Depp JL, Taylor JJ. et al. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. Embo J. 2003;22(10):2453–2462. [PMC free article: PMC155984] [PubMed: 12743039]
- 69.
- Hamada F, Bienz M. The APC tumor suppressor binds to C-terminal binding protein to divert nuclear beta-catenin from TCF. Dev Cell. 2004;7(5):677–685. [PubMed: 15525529]
- 70.
- Subramanian T, La Regina M, Chinnadurai G. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene. 1989;4(4):415–420. [PubMed: 2524023]
- 71.
- Subramanian T, Malstrom SE, Chinnadurai G. Requirement of the C-terminal region of adenovirus E1a for cell transformation in cooperation with E1b. Oncogene. 1991;6(7):1171–1173. [PubMed: 1830644]
- 72.
- Quinlan MP, Douglas JL. Immortalization of primary epithelial cells requires first- and second-exon functions of adenovirus type 5 12S. J Virol. 1992;66(4):2020–2030. [PMC free article: PMC288991] [PubMed: 1532211]
- 73.
- Gopalakrishnan S, Douglas JL, Quinlan MP. Immortalization of primary epithelial cells by E1A 12S requires late, second exon-encoded functions in addition to complex formation with pRB and p300. Cell Growth Differ. 1997;8(5):541–551. [PubMed: 9149905]
- 74.
- Glasspool RM, Burns S, Hoare SF. et al. The hTERT and hTERC telomerase gene promoters are activated by the second exon of the adenoviral protein, E1A, identifying the transcriptional corepressor CtBP as a potential repressor of both genes. Neoplasia. 2005;7(6):614–622. [PMC free article: PMC1501281] [PubMed: 16036112]
- 75.
- Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19(33):3823–3828. [PubMed: 10949939]
- 76.
- Johansson C, Zhao H, Bajak E. et al. Impact of the interaction between adenovirus E1A and CtBP on host cell gene expression. Virus Res. 2005 [PubMed: 15899534]
- 77.
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005 [PubMed: 16098727]
- 78.
- Alpatov R, Munguba GC, Caton P. et al. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol Cell Biol. 2004;24(23):10223–10235. [PMC free article: PMC529029] [PubMed: 15542832]
- 79.
- Zhang Q, Yoshimatsu Y, Hildebrand J. et al. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115(2):177–186. [PubMed: 14567915]
- 80.
- Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci USA. 2005;102(8):2802–2807. [PMC free article: PMC549470] [PubMed: 15708980]
- 81.
- Takahashi S, Licht JD. The human promyelocytic leukemia zinc finger gene is regulated by the Evi-1 oncoprotein and a novel guanine-rich site binding protein. Leukemia. 2002;16(9):1755–1762. [PubMed: 12200691]
- 82.
- Izutsu K, Kurokawa M, Imai Y. et al. The t(3;21) fusion product, AML1/Evi-1 blocks AML1-induced transactivation by recruiting CtBP. Oncogene. 2002;21(17):2695–2703. [PubMed: 11965542]
- 83.
- Kurokawa M, Mitani K, Irie K. et al. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394(6688):92–96. [PubMed: 9665135]
- 84.
- Izutsu K, Kurokawa M, Imai Y. et al. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood. 2001;97(9):2815–2822. [PubMed: 11313276]
- 85.
- Palmer S, Brouillet JP, Kilbey A. et al. Evi-1 transforming and repressor activities are mediated by CtBP corepressor proteins. J Biol Chem. 2001;276(28):25834–25840. [PubMed: 11328817]
- 86.
- Alliston T, Ko TC, Cao Y. et al. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem. 2005;280(25):24227–24237. [PubMed: 15849193]
- 87.
- Hickabottom M, Parker GA, Freemont P. et al. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the corepressor CtBP. J Biol Chem. 2002;7:7. [PubMed: 12372828]
- 88.
- Touitou R, Hickabottom M, Parker G. et al. Physical and functional interactions between the corepressor CtBP and the Epstein-barr virus nuclear antigen EBNA3C. J Virol. 2001;75(16):7749–7755. [PMC free article: PMC115013] [PubMed: 11462050]
- 89.
- Mirnezami AH, Campbell SJ, Darley M. et al. Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription. Curr Biol. 2003;13(14):1234–1239. [PubMed: 12867035]
- 90.
- Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: A dual mechanism. Genes Dev. 1997;11(15):1974–1986. [PMC free article: PMC316412] [PubMed: 9271120]
- 91.
- Bannister AJ, Kouzarides T. Histone methylation: Recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. [PubMed: 14975312]
- 92.
- Kuzmichev A, Nishioka K, Erdjument-Bromage H. et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. [PMC free article: PMC187479] [PubMed: 12435631]
- 93.
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17(15):1823–1828. [PMC free article: PMC196225] [PubMed: 12897052]
- 94.
- Fischle W, Wang Y, Jacobs SA. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–1881. [PMC free article: PMC196235] [PubMed: 12897054]
Publication Details
Author Information and Affiliations
Authors
G Chinnadurai*.Affiliations
Notes
Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Chinnadurai G. CtBP Family Proteins: Unique Transcriptional Regulators in the Nucleus with Diverse Cytosolic Functions. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.