Only those changes in DNA sequence that have functional consequences are known as disease-causing mutations. One such frequently occurring mutation causes a premature stop codon to appear in the middle of a protein-coding sequence of messenger RNA (mRNA). Stop codons (a triplet of nucleotides: UAA, UAG, or UGA) normally signal the end of the stretch of mRNA that is translated into protein so that when one appears early, the result can be a truncated protein that could have nasty consequences for the host organism.
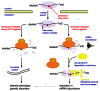
However, a mechanism known as "nonsense-mediated decay" has evolved to detect these harmful RNAs, and sequence analysis suggests that it may have been conserved in eukaryotic organisms, including humans. In yeast, three proteins have been identified that are required to seek and destroy the partly translated RNAs: Upf1p, Upf2p, and Upf3p.
Upf1p is an RNA unwinding enzyme, a helicase, that requires ATP for activity. Unfortunately, Upf1p will unwind pretty much anything, not just the problem mRNAs. So Upf2p and Upf3p are thought to be required to help Upf1p discriminate between nonsense and "real" mRNAs.
How do the core proteins work in synergy to trigger nonsense-mediated decay? One possibility is that Upf3p, along with several other ribonuclear proteins, may first bind to an mRNA as it is being exported from the nucleus en route to the ribosome, the site of protein synthesis. If the mRNA is fully translated into protein, Upf3p and the other protein factors are displaced. However, if there is a premature stop codon, Upf3p and cohorts may sit tight and mark the mutant mRNA as one that needs to be disposed of.
Experiments have shown that Upf3p can bind Upf2p. Once bound, Upf2p could signal to the "termination complex", a mixed bag of termination factors that includes Upf1p. This results in the release of the incomplete polypeptide from the ribosome, mRNA unwinding by Upf1p and, exposure of the mRNA for total degradation by exonuclease.
Although this model is attractive, more experiments are required to show that this actually happens in a living yeast cell.
Many of the mutations that form a premature stop codon lead to human disease, for example, those in BRCA1 that lead to breast cancer, or those in NF1 that lead to neurofibromatosis type 1, to name just two. There are two ways by which nonsense-mediated decay can play a role in the disease process. The first occurs when the machinery is functioning correctly: if mutant mRNAs are removed, then there will be a reduction in the amount of mRNA and protein available in the cell. The second is when a mutation occurs in the nonsense-mediated decay process itself, such as a mutation in RENT1, a human homolog of Upf1p, resulting in a population of truncated proteins, which could be harmful when targeted to their site of function.
Publication Details
Publication History
Created: October 13, 1999.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Dean L, McEntyre J, editors. Coffee Break: Tutorials for NCBI Tools [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1999-. RNA surveillance: watching the defectives: detecting premature stop codons in mRNA halts the production of dangerous truncated proteins. 1999 Oct 13.