By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002.

Molecular Biology of the Cell. 4th edition.
Show detailsIn principle, every step required for the process of gene expression could be controlled. Indeed, one can find examples of each type of regulation, although any one gene is likely to use only a few of them. Controls on the initiation of gene transcription are the predominant form of regulation for most genes. But other controls can act later in the pathway from DNA to protein to modulate the amount of gene product that is made. Although these posttranscriptional controls, which operate after RNA polymerase has bound to the gene's promoter and begun RNA synthesis, are less common than transcriptional control, for many genes they are crucial.
In the following sections, we consider the varieties of posttranscriptional regulation in temporal order, according to the sequence of events that might be experienced by an RNA molecule after its transcription has begun (Figure 7-87).
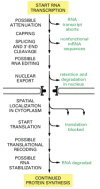
Figure 7-87
Possible post-transcriptional controls on gene expression. Only a few of these controls are likely to be important for any one gene.
Transcription Attenuation Causes the Premature Termination of Some RNA Molecules
In bacteria the expression of certain genes is inhibited by premature termination of transcription, a phenomenon called transcription attenuation. In some of these cases the nascent RNA chain adopts a structure that causes it to interact with the RNA polymerase in such a way as to abort its transcription. When the gene product is required, regulatory proteins bind to the nascent RNA chain and interfere with attenuation, allowing the transcription of a complete RNA molecule.
In eucaryotes transcription attenuation can occur by a number of distinct mechanisms. A well-studied example is found in HIV (the human AIDS virus). Once it has been integrated into the host genome, the viral DNA is transcribed by the cellular RNA polymerase II (see Figure 5-73). However, the host polymerase usually terminates transcription (for reasons that are not well-understood) after synthesizing transcripts of several hundred nucleotides and therefore does not efficiently transcribe the entire viral genome. When conditions for viral growth are optimal, this premature termination is prevented by a virus-encoded protein called Tat, which binds to a specific stem-loop structure in the nascent RNA that contains a “bulged base.” Once bound to this specific RNA structure (called Tar), Tat assembles several cellular proteins which allow the RNA polymerase to continue transcribing. The normal role of at least some of these cellular proteins is to prevent pausing and premature termination by RNA polymerase when it transcribes normal cellular genes. Eucaryotic genes often contain long introns; to transcribe a gene efficiently, RNA polymerase II cannot afford to linger at nucleotide sequences that happen to promote pausing. Thus a normal cellular mechanism has apparently been adapted by HIV to permit transcription of its genome to be controlled by a single viral protein.
Alternative RNA Splicing Can Produce Different Forms of a Protein from the Same Gene
As discussed in Chapter 6, the transcripts of many eucaryotic genes are shortened by RNA splicing, in which the intron sequences are removed from the mRNA precursor. We saw that a cell can splice the “primary transcript” in different ways and thereby make different polypeptide chains from the same gene—a process called alternative RNA splicing (see Figure 6-27 and Figure 7-88). A substantial proportion of higher eucaryotic genes (at least a third of human genes, it is estimated) produce multiple proteins in this way.
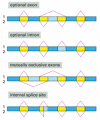
Figure 7-88
Four patterns of alternative RNA splicing. In each case a single type of RNA transcript is spliced in two alternative ways to produce two distinct mRNAs (1 and 2). The dark blue boxes mark exon sequences that are retained in both mRNAs. The light blue (more...)
When different splicing possibilities exist at several positions in the transcript, a single gene can produce dozens of different proteins. In one extreme case, a Drosophila gene may produce as many as 38,000 different proteins from a single gene through alternative splicing (Figure 7-89), although only a small fraction of these forms have thus far been experimentally observed. Considering that the Drosophila genome has approximately 14,000 identified genes, it is clear that the protein complexity of an organism can greatly exceed the number of its genes. This example also illustrates the perils in equating gene number with organism complexity. For example, alternative splicing is relatively rare in single-celled budding yeasts but very common in flies. Budding yeast has ~6200 genes, only 327 of which are subject to splicing, and nearly all of these have only a single intron. To say that flies have only 2–3 times as many genes as yeasts is to greatly underestimate the difference in complexity of these two genomes.
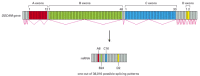
Figure 7-89
Alternative splicing of RNA transcripts of the Drosophila DSCAM gene. DSCAM proteins are axon guidance receptors that help to direct growth cones to their appropriate targets in the developing nervous system. The final mRNA contains 24 exons, four of (more...)
In some cases alternative RNA splicing occurs because there is an intron sequence ambiguity: the standard spliceosome mechanism for removing intron sequences (discussed in Chapter 6) is unable to distinguish cleanly between two or more alternative pairings of 5′ and 3′ splice sites, so that different choices are made by chance on different transcripts. Where such constitutive alternative splicing occurs, several versions of the protein encoded by the gene are made in all cells in which the gene is expressed.
In many cases, however, alternative RNA splicing is regulated rather than constitutive. In the simplest examples, regulated splicing is used to switch from the production of a nonfunctional protein to the production of a functional one. The transposase that catalyzes the transposition of the Drosophila P element, for example, is produced in a functional form in germ cells and a nonfunctional form in somatic cells of the fly, allowing the P element to spread throughout the genome of the fly without causing damage in somatic cells (see Figure 5-70). The difference in transposon activity has been traced to the presence of an intron sequence in the transposase RNA that is removed only in germ cells.
In addition to switching from the production of a functional protein to the production of a nonfunctional one, the regulation of RNA splicing can generate different versions of a protein in different cell types, according to the needs of the cell. Tropomyosin, for example, is produced in specialized forms in different types of cells (see Figure 6-27). Cell-type-specific forms of many other proteins are produced in the same way.
RNA splicing can be regulated either negatively, by a regulatory molecule that prevents the splicing machinery from gaining access to a particular splice site on the RNA, or positively, by a regulatory molecule that helps direct the splicing machinery to an otherwise overlooked splice site (Figure 7-90). In the case of the Drosophila transposase, the key splicing event is blocked in somatic cells by negative regulation.
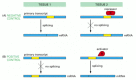
Figure 7-90
Negative and positive control of alternative RNA splicing. (A) Negative control, in which a repressor protein binds to the primary RNA transcript in tissue 2, thereby preventing the splicing machinery from removing an intron sequence. (B) Positive control, (more...)
Because of the plasticity of RNA splicing (see pp. 324–325), the blocking of a “strong” splicing site will often expose a “weak” site and result in a different pattern of splicing. Likewise, activating a suboptimal splice site can result in alternative splicing by suppressing a competing splice site. Thus the splicing of a pre-mRNA molecule can be thought of as a delicate balance between competing splice sites—a balance that can easily be tipped by regulatory proteins.
The Definition of a Gene Has Had to Be Modified Since the Discovery of Alternative RNA Splicing
The discovery that eucaryotic genes usually contain introns and that their coding sequences can be assembled in more than one way raised new questions about the definition of a gene. A gene was first clearly defined in molecular terms in the early 1940s from work on the biochemical genetics of the fungus Neurospora. Until then, a gene had been defined operationally as a region of the genome that segregates as a single unit during meiosis and gives rise to a definable phenotypic trait, such as a red or a white eye in Drosophila or a round or wrinkled seed in peas. The work on Neurospora showed that most genes correspond to a region of the genome that directs the synthesis of a single enzyme. This led to the hypothesis that one gene encodes one polypeptide chain. The hypothesis proved fruitful for subsequent research; as more was learned about the mechanism of gene expression in the 1960s, a gene became identified as that stretch of DNA that was transcribed into the RNA coding for a single polypeptide chain (or a single structural RNA such as a tRNA or an rRNA molecule). The discovery of split genes and introns in the late 1970s could be readily accommodated by the original definition of a gene, provided that a single polypeptide chain was specified by the RNA transcribed from any one DNA sequence. But it is now clear that many DNA sequences in higher eucaryotic cells can produce a set of distinct (but related) proteins by means of alternative RNA splicing. How then is a gene to be defined?
In those relatively rare cases in which two very different eucaryotic proteins are produced from a single transcription unit, the two proteins are considered to be produced by distinct genes that overlap on the chromosome. It seems unnecessarily complex, however, to consider most of the protein variants produced by alternative RNA splicing as being derived from overlapping genes. A more sensible alternative is to modify the original definition to count as a gene any DNA sequence that is transcribed as a single unit and encodes one set of closely related polypeptide chains (protein isoforms). This definition of a gene also accommodates those DNA sequences that encode protein variants produced by posttranscriptional processes other than RNA splicing, such as translational frameshifting (see Figure 6-78), regulated poly-A addition, and RNA editing (to be discussed below).
Sex Determination in Drosophila Depends on a Regulated Series of RNA Splicing Events
We now turn to one of the best understood examples of regulated RNA splicing. In Drosophila the primary signal for determining whether the fly develops as a male or female is the X chromosome/autosome ratio. Individuals with an X chromosome/autosome ratio of 1 (normally two X chromosomes and two sets of autosomes) develop as females, whereas those with a ratio of 0.5 (normally one X chromosome and two sets of autosomes) develop as males. This ratio is assessed early in development and is remembered thereafter by each cell. Three crucial gene products transmit information about this ratio to the many other genes that specify male and female characteristics (Figure 7-91). As explained in Figure 7-92, sex determination in Drosophila depends on a cascade of regulated RNA splicing events that involves these three gene products.
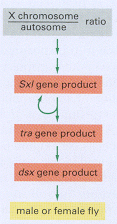
Figure 7-91
Sex determination in Drosophila. The gene products shown act in a sequential cascade to determine the sex of the fly according to the X chromosome/autosome ratio. The genes are called sex-lethal (Sxl), transformer (tra), and doublesex (dsx) because of (more...)
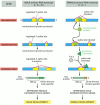
Figure 7-92
The cascade of changes in gene expression that determines the sex of a fly through alternative RNA splicing. An X chromosome/ autosome ratio of 0.5 results in male development. Male is the “default” pathway in which the Sxl and tra genes (more...)
Although Drosophila sex determination provides one of the best-understood examples of a regulatory cascade based on RNA splicing, it is not clear why the fly should use this strategy. Other organisms (the nematode, for example) use an entirely different scheme for sex determination—one based on transcriptional and translational controls. Moreover, the Drosophila male-determination pathway requires that a number of nonfunctional RNA molecules be continually produced, which seems unnecessarily wasteful. One speculation is that this RNA-splicing cascade exploits an ancient control device, left over from the early stage of evolution where RNA was the predominant biological molecule, and controls of gene expression would have had to be based almost entirely on RNA-RNA interactions (discussed in Chapter 6).
A Change in the Site of RNA Transcript Cleavage and Poly-A Addition Can Change the C-terminus of a Protein
We saw in Chapter 6 that the 3′ end of a eucaryotic mRNA molecule is not formed by the termination of RNA synthesis by the RNA polymerase. Instead, it results from an RNA cleavage reaction that is catalyzed by additional factors while the transcript is elongating (see Figure 6-37). A cell can control the site of this cleavage so as to change the C-terminus of the resultant protein.
A well-studied example is the switch from the synthesis of membrane-bound to secreted antibody molecules that occurs during the development of B lymphocytes. Early in the life history of a B lymphocyte, the antibody it produces is anchored in the plasma membrane, where it serves as a receptor for antigen. Antigen stimulation causes B lymphocytes to multiply and to begin secreting their antibody. The secreted form of the antibody is identical to the membrane-bound form except at the extreme C-terminus. In this part of the protein, the membrane-bound form has a long string of hydrophobic amino acids that traverses the lipid bilayer of the membrane, whereas the secreted form has a much shorter string of hydrophilic amino acids. The switch from membrane-bound to secreted antibody therefore requires a different nucleotide sequence at the 3′ end of the mRNA; this difference is generated through a change in the length of the primary RNA transcript, caused by a change in the site of RNA cleavage, as shown in Figure 7-93. This change is caused by an increase of the concentration of a subunit of CStF, the protein that binds to the G/U rich sequences of RNA cleavage and poly-A addition sites and promotes RNA cleavage (see Figures 6-37 and 6-38). The first cleavage-poly-A addition site encountered by an RNA polymerase transcribing the antibody gene is suboptimal and is usually skipped in unstimulated B lymphocytes, leading to production of the longer RNA transcript. When antibody stimulation causes an increase in CSTF concentration, cleavage now occurs at the suboptimal site, and the shorter transcript is produced. In this way a change in concentration of a general RNA processing factor can produce specific effects on a relatively small number of genes.

Figure 7-93
Regulation of the site of RNA cleavage and poly-A addition determines whether an antibody molecule is secreted or remains membrane-bound. In unstimulated B lymphocytes (left), a long RNA transcript is produced, and the intron sequence near its 3′ (more...)
RNA Editing Can Change the Meaning of the RNA Message
The molecular mechanisms used by cells are a continual source of surprises. An example is the process of RNA editing, which alters the nucleotide sequences of mRNA transcripts once they are transcribed. In Chapter 6, we saw that rRNAs and tRNAs are modified posttranscriptionally. In this section we see that some mRNAs are modified in ways that change the coded message they carry. The most dramatic form of RNA editing was discovered in RNA transcripts that code for proteins in the mitochondria of trypanosomes. Here, one or more U nucleotides are inserted (or, less frequently, removed) from selected regions of a transcript, causing major modifications in both the original reading frame and the sequence, thereby changing the meaning of the message. For some genes the editing is so extensive that over half of the nucleotides in the mature mRNA are U nucleotides that were inserted during the editing process. The information that specifies exactly how the initial RNA transcript is to be altered is contained in a set of 40- to 80-nucleotide-long RNA molecules that are transcribed separately. These so-called guide RNAs have a 5′ end that is complementary in sequence to one end of the region of the transcript to be edited; this is followed by a sequence that specifies the set of nucleotides to be inserted into the transcript, which is followed in turn by a continuous run of U nucleotides. The editing mechanism is remarkably complex, with the U nucleotides at the 3′ end of the guide RNA being transferred directly into the transcript, as illustrated in Figure 7-94.
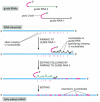
Figure 7-94
RNA editing in the mitochondria of trypanosomes. Guide RNAs contain at their 3′ end a stretch of poly U, which donates U nucleotides to sites on the RNA transcript that mispair with the guide RNA; thus the poly-U tail gets shorter as editing proceeds (more...)
Extensive editing of mRNA sequences has also been found in the mitochondria of many plants, with nearly every mRNA being edited to some extent. In this case, however, RNA bases are changed from C to U, without nucleotide insertions or deletions. Often many of the Cs in an mRNA are affected by editing, changing 10% or more of the amino acids that the mRNA encodes.
We can only speculate as to why the mitochondria of trypanosomes and plants make use of such extensive RNA editing. The suggestions that seem most reasonable are based on the premise that mitochondria contain a primitive genetic system that offers scanty opportunities for other forms of control. There is evidence that editing is regulated to produce different mRNAs under different conditions, so that RNA editing can be viewed as a primitive way to change the expression of genes, a relic, perhaps, of mechanisms that operated in very ancient cells, where most catalyses were probably carried out by RNA molecules rather than by proteins.
RNA editing of a much more limited kind occurs in mammals. One of the most important types is the enzymatic deamination of adenine to produce inosine (see Figure 6-55), which occurs at selected positions in some pre-mRNAs. In some cases, this modification changes the splicing pattern of the RNA; in others, it changes the meanings of codons. Because inosine base pairs with cytosine, A-to-I editing can result in a protein with an altered amino acid sequence. This editing is carried out by protein enzymes called ADARs (adenosine deaminases acting on RNA); these enzymes recognize a double-stranded RNA structure that is formed by base pairing between the site to be edited and a complementary sequence located elsewhere on the same RNA molecule, typically in a 3′ intron (Figure 7-95). An especially important example of A-to-I editing takes place in the pre-mRNA that codes for a transmitter-gated ion channel in the brain. A single edit changes a glutamine to an arginine; the affected amino acid lies on the inner wall of the channel, and the editing change alters the Ca2+ permeability of the channel. The importance of this edit in mice has been demonstrated by deleting the relevant ADAR gene. The mutant mice are prone to epileptic seizures and die during or shortly after weaning. If the gene for the gated ion channel is mutated to produce the edited form of the protein directly, mice lacking the ADAR develop normally, showing that editing of the ion channel RNA is normally crucial for proper brain development. Mice and humans have several additional ADAR genes, and deletion of one of these in mice causes death of the mouse embryo before birth. Because these mice have severe defects in the production of red blood cell precursors, it is likely that RNA editing is also essential for the proper development of the hemopoietic system.
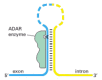
Figure 7-95
Mechanism of A-to-I RNA editing in mammals. The position of an edit is signaled by RNA sequences carried on the same RNA molecule. Typically, a sequence complementary to the position of the edit is present in an intron, and the resulting double-stranded (more...)
C-to-U editing has also been observed in mammals. In one example, that of the apolipoprotein-B mRNA, a C to U change creates a stop codon that causes a truncated version of this large protein to be made in a tissue-specific manner. Why editing in mammalian cells exists at all is a mystery. One idea is that it arose in evolution to correct “mistakes” in the genome. Another is that is provides yet another way for the cell to produce a variety of related proteins from a single gene. A third view is that editing is merely one of a large number of haphazard, makeshift devices that have originated through random mutation and have been perpetuated because they happen to contribute to a useful effect.
RNA Transport from the Nucleus Can Be Regulated
It has been estimated that in mammals only about one-twentieth of the total mass of RNA synthesized ever leaves the nucleus. We saw in Chapter 6 that most mammalian RNA molecules undergo extensive processing and the “left-over” RNA fragments (excised introns and RNA sequences 3′ to the cleavage/poly-A site) are degraded in the nucleus. Incompletely processed and otherwise damaged RNAs are also eventually degraded in the nucleus as part of the quality control system of RNA production. This degradation is carried out by the exosome, a large protein complex that contains, as subunits, several different RNA exonucleases.
As described in Chapter 6, the export of RNA molecules from the nucleus is delayed until processing has been completed. Therefore any mechanism that prevents the completion of RNA splicing on a particular RNA molecule could in principle block the exit of that RNA from the nucleus. This feature forms the basis for one of the best understood examples of regulated nuclear transport of mRNA, which occurs in HIV, the virus that causes AIDS.
HIV is a retrovirus—an RNA virus that, once inside a cell, directs the formation of a double-stranded DNA copy of its genome which is then inserted into the host genome (see Figure 5-73). Once inserted, the viral DNA is transcribed as one long RNA molecule by the host cell RNA polymerase II. This transcript is then spliced in many different ways to produce over 30 different species of mRNA, which in turn, are translated into a variety of different proteins (Figure 7-96). In order to make progeny virus, entire, unspliced viral transcripts must be exported from the nucleus to the cytosol where they are packaged into viral capsids (see Figure 5-73). Moreover, several of the HIV mRNAs are alternatively spliced in such a way that they still carry complete introns. The host cell's block to the nuclear export of unspliced RNA (and its subsequent degradation) therefore presents a special problem for HIV, and it is overcome in an ingenious way.
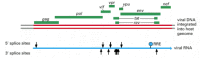
Figure 7-96
The compact genome of HIV, the human AIDS virus. The positions of the nine HIV genes are shown in green. The red double line indicates a DNA copy of the viral genome which has become integrated into the host DNA (gray). Note that the coding regions of (more...)
The virus encodes a protein (called Rev) that binds to a specific RNA sequence (called the Rev responsive element, RRE) located within a viral intron. The Rev protein interacts with a nuclear export receptor (exportin 1), which directs the movement of viral RNAs through nuclear pores into the cytosol despite the presence of intron sequences. We discuss in detail the way that export receptors function in Chapter 12.
The regulation of nuclear export by Rev has several important consequences for HIV growth and pathogenesis. In addition to ensuring the nuclear export of specific unspliced RNAs, it divides the viral infection into an early phase (where Rev is translated from a fully spliced RNA and RNAs containing an intron are retained in the nucleus and degraded) and a late phase (where unspliced RNAs are exported due to Rev function). This timing helps the virus replicate by providing the gene products roughly in the order in which they are needed (Figure 7-97). It is also possible that regulation by Rev helps the HIV virus to achieve latency, a condition where the HIV genome has become integrated into the host cell genome but the production of viral proteins has temporarily ceased. If, after its initial entry into a host cell, conditions became unfavorable for viral transcription and replication, Rev is made at levels too low to promote export of unspliced RNA. This situation stalls the viral growth cycle. When conditions for viral replication improve, Rev levels increase, and the virus can enter the replication cycle.
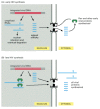
Figure 7-97
Regulation of nuclear export by the HIV Rev protein. Early in HIV infection (A), only the fully spliced RNAs (which contain the coding sequences for Rev, Tat, and Nef) are exported from the nucleus and translated. Once sufficient Rev protein has accumulated (more...)
Some mRNAs Are Localized to Specific Regions of the Cytoplasm
Once a newly made eucaryotic mRNA molecule has passed through a nuclear pore and entered the cytosol, it is typically met by ribosomes, which translate it into a polypeptide chain (see Figure 6-40). If the mRNA encodes a protein that is destined to be secreted or expressed on the cell surface, it will be directed to the endoplasmic reticulum (ER) by a signal sequence at the protein's amino terminus; components of the cell's protein-sorting apparatus recognize the signal sequence as soon as it emerges from the ribosome and direct the entire complex of ribosome, mRNA, and nascent protein to the membrane of the ER, where the remainder of the polypeptide chain is synthesized, as discussed in Chapter 12. In other cases the entire protein is synthesized by free ribosomes in the cytosol, and signals in the completed polypeptide chain may then direct the protein to other sites in the cell.
Some mRNAs are themselves directed to specific intracellular locations before translation begins. Presumably it is advantageous for the cell to position its mRNAs close to the sites where the protein produced from the mRNA is required. The signals that direct mRNA localization are typically located in the 3′ untranslated region (UTR) of the mRNA molecule—a region that extends from the stop codon, which terminates protein synthesis, to the start of the poly-A tail (see Figure 6-22A). A striking example of mRNA localization is seen in the Drosophila egg, where the mRNA encoding the bicoid gene regulatory protein is localized by attachment to the cytoskeleton at the anterior tip of the developing egg. When the translation of this mRNA is triggered by fertilization, a gradient of the bicoid protein is generated that plays a crucial part in directing the development of the anterior part of the embryo (shown in Figure 7-52 and discussed in more detail in Chapter 21). Many mRNAs in somatic cells are localized in a similar way. The mRNA that encodes actin, for example, is localized to the actin-filament-rich cell cortex in mammalian fibroblasts by means of a 3′ UTR signal.
RNA localization has been observed in many organisms, including unicellular fungi, plants, and animals, and it is likely to be a common mechanism used by cells to concentrate high-level production of proteins at specific sites. Several distinct mechanisms for mRNA localization have been discovered (Figure 7-98), but all of them require specific signals in the mRNA itself, usually concentrated in the 3′ UTR (Figure 7-99).
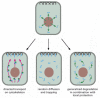
Figure 7-98
Three mechanisms for the localization of mRNAs. The mRNA to be localized leaves the nucleus through nuclear pores (top). Some localized mRNAs (left diagram) travel to their destination by associating with cytoskeleton motors (green). As described in Chapter (more...)
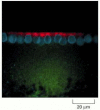
Figure 7-99
The importance of the 3′ UTR in localizing mRNAs to specific regions of the cytoplasm. For this experiment, two different fluorescently-labeled RNAs were prepared by transcribing DNA in vitro in the presence of fluorescently-labeled derivatives (more...)
We saw in Chapter 6 that the 5′ cap and the 3′ poly-A tail are necessary for efficient translation, and their presence on the same mRNA molecule thereby signals to the translation machinery that the mRNA molecule is intact. As just described, the 3′ UTR often contains a “zip code,” which directs mRNAs to different places in the cell. In this chapter, we will also see that mRNAs also carry information specifying the average length of time each mRNA persists in the cytosol and the efficiency with which each mRNA is translated into protein. In a broad sense, the untranslated regions of eucaryotic mRNAs resemble the transcriptional control regions of genes: their nucleotide sequences contain information specifying the way the RNA is to be used, and proteins that interpret this information bind specifically to these sequences. Thus, over and above the specification of the amino acid sequences of proteins, mRNA molecules are rich with many additional types of information.
Proteins That Bind to the 5′ and 3′ Untranslated Regions of mRNAs Mediate Negative Translational Control
Once an mRNA has been synthesized, one of the most common ways of regulating the levels of its protein product is by controlling the step in which translation is initiated. Even though the mechanistic details of translation initiation differ between eucaryotes and bacteria (as we saw in Chapter 6), some of the same basic regulatory strategies are used.
In bacterial mRNAs a conserved stretch of six nucleotides, the Shine-Dalgarno sequence, is always found a few nucleotides upstream of the initiating AUG codon. This sequence forms base pairs with the 16S RNA in the small ribosomal subunit, correctly positioning the initiating AUG codon in the ribosome. Because this interaction makes a major contribution to the efficiency of initiation, it provides the bacterial cell with a simple way to regulate protein synthesis through negative translational control mechanisms. These mechanisms generally involve blocking the Shine-Dalgarno sequence, either by covering it with a bound protein or by incorporating it into a base-paired region in the mRNA molecule. Many bacterial mRNAs have specific translational repressor proteins that can bind in the vicinity of the Shine-Dalgarno sequence and thereby inhibit translation of only that species of mRNA. For example, some ribosomal proteins can repress translation of their own mRNAs by binding to the 5′ untranslated region. This mechanism comes into play only when the ribosomal proteins are produced in excess over ribosomal RNA and are therefore not incorporated into ribosomes. In this way, it allows the cell to maintain correctly balanced quantities of the various components needed to form ribosomes. It is not hard to guess how this mechanism might have evolved. Ribosomal proteins assemble into ribosomes by binding to specific sites in rRNA; ingeniously, some of them exploit this RNA-binding ability to regulate their own production by binding to similar sites present in their own mRNAs.
Eucaryotic mRNAs do not contain a Shine-Dalgarno sequence. Instead, as discussed in Chapter 6, the selection of an AUG codon as a translation start site is largely determined by its proximity to the cap at the 5′ end of the mRNA molecule, which is the site at which the small ribosomal subunit binds to the mRNA and begins scanning for an initiating AUG codon. Despite the differences in translation initiation, eucaryotes also utilize translational repressors. Some bind to the 5′ end of the mRNA and thereby inhibit translation initiation. Others recognize nucleotide sequences in the 3′ UTR of specific mRNAs and decrease translation initiation by interfering with the communication between the 5′ cap and 3′ poly-A tail, which is required for efficient translation (see Figure 6-71).
A well-studied form of negative translational control in eucaryotes allows the synthesis of the intracellular iron storage protein ferritin to be increased rapidly if the level of soluble iron atoms in the cytosol rises. The iron regulation depends on a sequence of about 30 nucleotides in the 5′ leader of the ferritin mRNA molecule. This iron-response element folds into a stem-loop structure that binds a translation repressor protein called aconitase, which blocks the translation of any RNA sequence downstream (Figure 7-100). Aconitase is an iron-binding protein, and exposure of the cell to iron causes it to dissociate from the ferritin mRNA, releasing the block to translation and increasing the production of ferritin by as much as hundredfold.
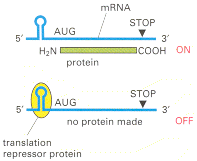
Figure 7-100
Negative translational control. This form of control is mediated by a sequence-specific RNA-binding protein that acts as a translation repressor. Binding of the protein to an mRNA molecule decreases the translation of the mRNA. Several cases of this type (more...)
The Phosphorylation of an Initiation Factor Globally Regulates Protein Synthesis
Eucaryotic cells decrease their overall rate of protein synthesis in response to a variety of situations, including deprivation of growth factors or nutrients, infection by viruses, and sudden increases in temperature. Much of this decrease is caused by the phosphorylation of the translation initiation factor eIF-2 by specific protein kinases that respond to the changes in conditions.
The normal function of eIF-2 was outlined in Chapter 6. It forms a complex with GTP and mediates the binding of the methionyl initiator tRNA to the small ribosomal subunit, which then binds to the 5′ end of the mRNA and begins scanning along the mRNA. When an AUG codon is recognized, the bound GTP is hydrolyzed to GDP by the eIF-2 protein, causing a conformational change in the protein and releasing it from the small ribosomal subunit. The large ribosomal subunit then joins the small one to form a complete ribosome that begins protein synthesis (see Figure 6-71).
Because eIF-2 binds very tightly to GDP, a guanine nucleotide exchange factor (see Figure 15-54), designated eIF-2B, is required to cause GDP release so that a new GTP molecule can bind and eIF-2 can be reused (Figure 7-101A). The reuse of eIF-2 is inhibited when it is phosphorylated—the phosphorylated eIF-2 binds to eIF-2B unusually tightly, inactivating eIF-2B. There is more eIF-2 than eIF-2B in cells, and even a fraction of phosphorylated eIF-2 can trap nearly all of the eIF-2B. This prevents the reuse of the nonphosphorylated eIF-2 and greatly slows protein synthesis (Figure 7-101B).
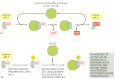
Figure 7-101
The elF-2 cycle. (A) The recycling of used elF-2 by a guanine nucleotide exchange factor (elF-2B). (B) elF-2 phosphorylation controls protein synthesis rates by tying up elF-2B.
Regulation of the level of active eIF-2 is especially important in mammalian cells, being part of the mechanism that allows them to enter a nonproliferating, resting state (called G0)—in which the rate of total protein synthesis is reduced to about one-fifth the rate in proliferating cells (discussed in Chapter 17).
Initiation at AUG Codons Upstream of the Translation Start Can Regulate Eucaryotic Translation Initiation
We saw in Chapter 6 that eucaryotic translation typically begins at the first AUG downstream of the 5′ end of the mRNA, as it is the first AUG encountered by a scanning small ribosomal subunit. But the nucleotides immediately surrounding the AUG also influence the efficiency of translation initiation. If the recognition site is poor enough, scanning ribosomal subunits will ignore the first AUG codon in the mRNA and skip to the second or third AUG codon instead. This phenomenon, known as “leaky scanning,” is a strategy frequently used to produce two or more closely related proteins, differing only in their amino termini, from the same mRNA. For example, it allows some genes to produce the same protein with and without a signal sequence attached at its amino terminus so that the protein is directed to two different locations in the cell. In some cases, the cell can regulate the relative abundance of the protein isoforms produced by leaky scanning; for example, a cell-type specific increase in the abundance of the initiation factor eIF-4F favors usage of the AUG closest to the 5′ end of the mRNA.
Another type of control found in eucaryotes uses one or more short open reading frames that lie between the 5′ end of the mRNA and the beginning of the gene. Often, the amino acid sequences coded by these upstream open reading frames (uORFs) are not critical; rather the uORFs serve a purely regulatory function. An uORF present on an mRNA molecule will generally decrease translation of the downstream gene by trapping a scanning ribosome initiation complex and causing the ribosome to translate the uORF and dissociate from the mRNA before it reaches the protein coding sequences.
When the activity of a general translation factor, such as eIF-2 (discussed above), is reduced, one might expect that the translation of all mRNAs would be reduced equally. Contrary to this expectation, however, the phosphorylation of eIF-2 can have selective effects, even enhancing the translation of specific mRNAs that contain uORFs. This can enable yeast cells, for example, to adapt to starvation for specific nutrients by shutting down the synthesis of all proteins except those that are required for synthesis of the nutrients that are missing. The details of this mechanism have been worked out for a specific yeast mRNA that encodes a protein called Gcn4, a gene regulatory protein that is required for the activation of many genes encoding proteins that are important for amino acid synthesis.
The GCN4 mRNA contains four short uORFs, and these are responsible for selectively increasing the translation of GCN4 in response to eIF-2 phosphorylation provoked by amino acid starvation. The mechanism by which GCN4 translation is increased is complex. In outline, ribosomal subunits move along the mRNA, encountering each of the uORFs but translating only a subset of them; if the fourth uORF is translated, as is the case in unstarved cells, the ribosomes dissociate at the end of the uORF, and translation of GCN4 is inefficient. The decrease in eIF-2 activity makes it more likely that a scanning ribosome will move through the fourth uORF before it acquires the ability to initiate translation. Such a ribosome can then efficiently initiate translation on the GCN4 sequences, leading to the production of proteins that promote amino acid synthesis inside the cell.
Internal Ribosome Entry Sites Provide Opportunities for Translation Control
Although approximately 90% of eucaryotic mRNAs are translated beginning with the first AUG downstream from the 5′ cap, certain AUGs, as we saw in the last section, can be skipped over during the scanning process. In this section, we discuss yet another way that cells can initiate translation at positions distant from the 5′ end of the mRNA. In these cases, translation is initiated directly at specialized RNA sequences, each of which is called an internal ribosome entry site (IRES). An IRES can occur in many different places in an mRNA. In some unusual cases, two distinct protein coding sequences are carried in tandem on the same eucaryotic mRNA; translation of the first occurs by the usual scanning mechanism and translation of the second through an IRES. IRESs are typically several hundred nucleotides in length and fold into specific structures that bind many, but not all, of the same proteins that are used to initiate normal cap-dependent translation (Figure 7-102). In fact, different IRESs require different subsets of initiation factors. However, all of them bypass the need for a 5′ cap structure and the translation initiation factor that recognizes it, eIF-4E.

Figure 7-102
Two mechanisms of translation initiation. (A) The cap-dependent mechanism requires a set of initiation factors whose assembly on the mRNA is stimulated by the presence of a 5′ cap and a poly-A tail (see also Figure 6-71). (B) The IRES-dependent (more...)
IRESs were first discovered in certain mammalian viruses where they provide a clever way for the virus to take over its host cell's translation machinery. On infection, these viruses produce a protease (encoded in the viral genome) that cleaves the cellular translation factor eIF-4G and thereby renders it unable to bind to eIF-4E, the cap-binding complex. This shuts down the great majority of host cell translation and effectively diverts the translation machinery to the IRES sequences, which are present on many viral mRNAs. The truncated eIF-4G remains competent to initiate translation at these internal sites and may even stimulate the translation of certain IRES-containing viral mRNAs.
The selective activation of IRES-mediated translation also occurs on cellular mRNAs. For example, when eucaryotic cells enter M phase of the cell cycle, the overall rate of translation drops to approximately 25% that in interphase cells. This drop is largely caused by a cell-cycle dependent dephosphorylation of the cap binding complex , eIF-4E, which lowers its affinity for the 5′ cap. IRES-containing mRNAs, however, are immune to this effect, and their relative translation rates therefore increase as the cell enters M phase.
Finally, when mammalian cells enter the programmed cell death pathway (discussed in Chapter 17), eIF-4G is cleaved, and a general decrease in translation ensues. Some proteins critical for the control of cell death are translated from IRES-containing mRNAs, and they continue to be synthesized. It seems that one of the main advantages of the IRES mechanism for the cell is that it allows selected mRNAs to be translated at a high rate despite a general decrease in the cell's capacity to initiate protein synthesis.
Gene Expression Can Be Controlled By a Change In mRNA Stability
The vast majority of mRNAs in a bacterial cell are very unstable, having a half-life of about 3 minutes. Exonucleases, which degrade in the 3′ to 5′ direction, are usually responsible for the rapid destruction of these mRNAs. Because its mRNAs are both rapidly synthesized and rapidly degraded, a bacterium can adapt quickly to environmental changes.
The mRNAs in eucaryotic cells are more stable. Some, such as that encoding β-globin, have half-lives of more than 10 hours. Many, however, have half-lives of 30 minutes or less. These unstable mRNAs often code for regulatory proteins, such as growth factors and gene regulatory proteins, whose production rates need to change rapidly in cells.
Two major degradation pathways exist for eucaryotic mRNAs, and sequences in each mRNA molecule determine the pathway and kinetics of degradation. The most common pathway involves the gradual shortening of the poly-A tail. We saw in Chapter 6 that capping and polyadenylation of mRNA molecules occurs in the nucleus. Once in the cytosol, the poly-A tails (which average about 200 As in length) are gradually shortened by an exonuclease that chews away the tail in the 3′ to 5′ direction. Once a critical threshold of tail shortening has been reached (approximately 30 A's remaining), the 5′ cap is removed (a process called “decapping”), and the RNA is rapidly degraded (Figure 7-103A).

Figure 7-103
Two mechanisms of eucaryotic mRNA decay. (A) Deadenylation-dependent decay. Most eucaryotic mRNAs are degraded by this pathway. The critical threshold of poly-A tail length that induces decay may correspond to the loss of the poly-A binding proteins (see (more...)
Nearly all mRNAs are subject to poly-A tail shortening, decapping, and eventual degradation, but the rate at which this occurs differs from one species of mRNA to the next. The proteins that carry out tail-shortening compete directly with the machinery that catalyzes translation; therefore, any factors that affect the translation efficiency of an mRNA will tend to have the opposite effect on its degradation (Figure 7-104). In addition, many mRNAs carry in their 3′ UTR sequences binding sites for specific proteins that increase or decrease the rate of poly-A tail shortening. For example, many unstable mRNAs contain stretches of AU sequences, which greatly enhance the shortening rate.
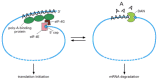
Figure 7-104
The competition between mRNA translation and mRNA decay. The same two features of mRNA—the 5′ cap and the 3′ poly-A site—are used in both translation initiation and deadenylation-dependent mRNA decay (see Figure 7-103). (more...)
A second pathway by which mRNA is degraded begins with the action of specific endonucleases, which simply cleave the poly-A tail from the rest of the mRNA in one step (see Figure 7-103). The mRNAs that are degraded in this way carry specific nucleotide sequences, typically in their 3′ UTR, that serve as recognition sequences for the endonucleases.
The stability of an mRNA can be changed in response to extracellular signals. For example, the addition of iron to cells decreases the stability of the mRNA that encodes the receptor protein that binds the iron-transporting protein transferrin, causing less of this receptor to be made. Interestingly, this effect is mediated by the iron-sensitive RNA-binding protein aconitase, which, as we discussed above, also controls ferritin mRNA translation. Aconitase can bind to the 3′ UTR of the transferrin receptor mRNA and cause an increase in receptor production by blocking endonucleolytic cleavage of the mRNA. On the addition of iron, aconitase is released from the mRNA, decreasing mRNA stability (Figure 7-105).
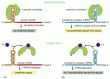
Figure 7-105
Two posttranslational controls mediated by iron. In response to an increase in iron concentration in the cytosol, a cell increases its synthesis of ferritin in order to bind the extra iron (A) and decreases its synthesis of transferrin receptors in order (more...)
Cytoplasmic Poly-A Addition Can Regulate Translation
The initial polyadenylation of an RNA molecule (discussed in Chapter 6) occurs in the nucleus, apparently automatically for nearly all eucaryotic mRNA precursors. As we have just seen, the poly-A tails on most mRNAs gradually shorten in the cytosol, and the RNAs are eventually degraded. In some cases, however, the poly-A tails of specific mRNAs are lengthened in the cytosol, and this mechanism provides an additional form of translational regulation.
Maturing oocytes and eggs provide the most striking example. Many of the normal mRNA degradation pathways seem to be disabled in these giant cells, so that the cells can build up large stores of mRNAs in preparation for fertilization. Many mRNAs are stored in the cytoplasm with only 10 to 30 As at their 3′ end, and in this form they are not translated. At specific times during oocyte maturation and postfertilization, when the proteins encoded by these mRNAs are required, poly A is added to selected mRNAs, greatly stimulating the initiation of their translation.
Nonsense-mediated mRNA Decay Is Used as an mRNA Surveillance System in Eucaryotes
We saw in Chapter 6 that mRNA production in eucaryotes occurs by an elaborately choreographed series of synthesis and processing steps. Only when all of the steps of mRNA production have been completed are the mRNAs exported from the nucleus to the cytosol for translation into protein. If any of those steps go awry, the RNA is eventually degraded in the nucleus (along with excised introns) by the exosome, a large protein complex that contains at least ten 3′-to-5′ RNA exonucleases. The eucaryotic cell has an additional mechanism, called nonsense-mediated mRNA decay, that eliminates certain types of aberrant mRNAs before they can be efficiently translated into protein. This mechanism was discovered when mRNAs that contain misplaced in-frame translation stop codons (UAA, UAG, or UGA) were found to be rapidly degraded. These stop codons can arise either from mutation or from incomplete splicing: in both cases, the phenomenon was observed. This mRNA surveillance system therefore prevents the synthesis of abnormally truncated proteins which, as we have seen, can be especially dangerous to the cell. But how are these potentially harmful mRNAs recognized by the cell?
In vertebrates, the critical feature of mRNA that is sensed by the nonsense-mediated decay system is the spatial relationship between the first in-frame termination codon and the exon-exon boundaries formed by RNA splicing. If the stop codon lies downstream (3′) of all the exon-exon boundaries, the mRNA is spared from nonsense-mediated decay; if, on the other hand, a stop codon is located upstream (5′) to an exon-exon boundary, the mRNA is degraded. Translating ribosomes, in conjunction with other surveillance proteins, assess this relationship for each individual mRNA. Exactly how this is accomplished is not understood in detail, but it is easy to understand why ribosomes must play a part: only in-frame termination codons trigger nonsense-mediated decay, and it is the relationship between the ribosome and the mRNA that defines the reading frame. According to one model (Figure 7-106), proteins in the nucleus bind to and thereby mark the exon-exon junctions following RNA splicing. As the mRNA leaves the nucleus, it remains at the nuclear periphery and is joined by a set of additional surveillance proteins as translation begins. The first round of translation of an individual mRNA molecule would, in this view, be used simply to assess the fitness of the mRNA for further rounds of translation. If the mRNA passes this test, translation begins in earnest as the mRNA is released to diffuse through the cytosol.
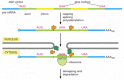
Figure 7-106
A model for nonsense-mediated mRNA decay. According to this model, nuclear proteins (orange) mark the exon-exon boundaries on a spliced mRNA molecule. These proteins are thought to assemble in concert with the splicing reaction and may also be involved (more...)
Nonsense-mediated decay may have been especially important in evolution, allowing eucaryotic cells to more easily explore new genes formed by DNA rearrangements, mutations, or alternative patterns of splicing—by selecting only those mRNAs for translation that produce a full-length protein. Nonsense-mediated decay is also important in cells of the developing immune system, where the extensive DNA rearrangements that occur (see Figure 24-37) often generate premature termination codons. The mRNAs produced from such rearranged genes are degraded by this surveillance system, thereby avoiding the toxic effects of truncated proteins.
RNA Interference Is Used by Cells to Silence Gene Expression
Eucaryotic cells use a specialized type of RNA degradation as a defense mechanism to destroy foreign RNA molecules, specifically those that can be identified by virtue of their occurrence within the cell in double-stranded form. Termed RNA interference (RNAi), this mechanism is found in a wide variety of organisms, including single-celled fungi, plants, worms, mice, and probably humans—suggesting that it is an evolutionarily ancient defense mechanism. In plants, RNA interference protects cells against RNA viruses. In other types of organisms, it is thought to protect against the proliferation of transposable elements that replicate via RNA intermediates (see Figure 5-76). Many transposable elements and plant viruses produce double-stranded RNA, at least transiently, in their life cycles. RNAi not only helps to keep such infestations in check, but also provides scientists with a powerful experimental technique to turn off the expression of individual cellular genes (see Figure 8-65).
The presence of free, double-stranded RNA triggers RNAi by attracting a protein complex containing an RNA nuclease and an RNA helicase. This protein complex cleaves the double-stranded RNA into small (approximately 23 nucleotide pair) fragments which remain associated with the enzyme. The bound RNA fragments then direct the enzyme complex to other RNA molecules that have complementary nucleotide sequences, and the enzyme degrades these as well. These other molecules can be either single- or double-stranded (as long as they have a complementary strand). In this way, the experimental introduction of a double-stranded RNA molecule can be used by scientists to inactivate specific cellular mRNAs (Figure 7-107).

Figure 7-107
The mechanism of RNA interference. On the left is shown the fate of foreign double-stranded RNA molecules. They are recognized by an RNase, present in a large protein complex, and degraded into short fragments that are approximately 23 nucleotide pairs (more...)
Each time it cleaves a new RNA, the enzyme complex is regenerated with a short RNA molecule, so that an original double-stranded RNA molecule can act catalytically to destroy many complementary RNAs. In addition, the short double-stranded RNA cleavage products themselves can be replicated by additional cellular enzymes, providing an even greater amplification of RNA interference activity (see Figure 7-107). This amplification ensures that once initiated, RNA interference can continue even after all the initiating double-stranded RNA has been degraded or diluted out. For example, it permits progeny cells to continue carrying out RNA interference that was provoked in the parent cells. In addition, the RNA interference activity can be spread by the transfer of RNA fragments from cell to cell. This is particularly important in plants (whose cells are linked by fine connecting channels, as discussed in Chapter 19), because it allows an entire plant to become resistant to an RNA virus after only a few of its cells have been infected.
Summary
Many steps in the pathway from RNA to protein are regulated by cells to control gene expression. Most genes are thought to be regulated at multiple levels, although control of the initiation of transcription (transcriptional control) usually predominates. Some genes, however, are transcribed at a constant level and turned on and off solely by posttranscriptional regulatory processes. These processes include (1) attenuation of the RNA transcript by its premature termination, (2) alternative RNA splice-site selection, (3) control of 3 ′ -end formation by cleavage and poly-A addition, (4) RNA editing, (5) control of transport from the nucleus to the cytosol, (6) localization of mRNAs to particular parts of the cell, (7) control of translation initiation, and (8) regulated mRNA degradation. Most of these control processes require the recognition of specific sequences or structures in the RNA molecule being regulated. This recognition is accomplished by either a regulatory protein or a regulatory RNA molecule.
- Transcription Attenuation Causes the Premature Termination of Some RNA Molecules
- Alternative RNA Splicing Can Produce Different Forms of a Protein from the Same Gene
- The Definition of a Gene Has Had to Be Modified Since the Discovery of Alternative RNA Splicing
- Sex Determination in Drosophila Depends on a Regulated Series of RNA Splicing Events
- A Change in the Site of RNA Transcript Cleavage and Poly-A Addition Can Change the C-terminus of a Protein
- RNA Editing Can Change the Meaning of the RNA Message
- RNA Transport from the Nucleus Can Be Regulated
- Some mRNAs Are Localized to Specific Regions of the Cytoplasm
- Proteins That Bind to the 5′ and 3′ Untranslated Regions of mRNAs Mediate Negative Translational Control
- The Phosphorylation of an Initiation Factor Globally Regulates Protein Synthesis
- Initiation at AUG Codons Upstream of the Translation Start Can Regulate Eucaryotic Translation Initiation
- Internal Ribosome Entry Sites Provide Opportunities for Translation Control
- Gene Expression Can Be Controlled By a Change In mRNA Stability
- Cytoplasmic Poly-A Addition Can Regulate Translation
- Nonsense-mediated mRNA Decay Is Used as an mRNA Surveillance System in Eucaryotes
- RNA Interference Is Used by Cells to Silence Gene Expression
- Summary
- Posttranscriptional Controls - Molecular Biology of the CellPosttranscriptional Controls - Molecular Biology of the Cell
Your browsing activity is empty.
Activity recording is turned off.
See more...