All Assay Guidance Manual content, except where otherwise noted, is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported license (CC BY-NC-SA 3.0), which permits copying, distribution, transmission, and adaptation of the work, provided the original work is properly cited and not used for commercial purposes. Any altered, transformed, or adapted form of the work may only be distributed under the same or similar license to this one.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Markossian S, Grossman A, Arkin M, et al., editors. Assay Guidance Manual [Internet]. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004-.
Abstract
Kinases are important drug targets that control cell growth, proliferation, differentiation and metabolism. This process involves phosphorylation of multiple substrates in cellular signal transduction pathways. Here, the authors provide a synopsis on assay technologies, mechanistic considerations for assay design and development pertaining to kinase enzymes. This chapter, along with some of the earlier chapters in this manual on basics of enzyme assays, mechanism of action and purity and identity considerations, serves as an excellent resource for beginners in HTS applications.
Introduction
Enzymatic phosphate transfers are one of the predominant mechanisms for regulating the growth, differentiation and metabolism of cells. The post-translational modification of proteins with phosphate leads to dramatic changes in conformation resulting in the modulation of binding, catalysis and recruitment of effector molecules that regulate cellular signaling pathways. Examples include the recruitment of SH2 domain containing proteins, the activation of gene transcription pathways and the activation or deactivation of specific cell surface receptors. The practical design and implementation of these assays for drug discovery and development applications will be the focus of this section.
The keys to protein kinase assay development lie in the ability to 1) choose an appropriate “readout” technology, 2) have ample quantities of enzymes, cell lines, antibodies and reference compounds, and 3) optimize the assay for buffer conditions, reagent concentrations, timing, stopping, order of addition, plate type and assay volume. The readout technologies present many options for assay development and often depend on the laboratory infrastructure, the cost of reagents, the desired substrate and the secondary assays needed to validate the compounds and determine the structure activity relationships (Table 1). In the end, they all require the measurement of photons emitted from the assay well in a microtiter plate. They differ in how the photons are generated, and what property (ie. wavelength or polarity) of the photons are measured.
Protein kinase enzyme assays require the co-factors ATP, magnesium (and sometimes manganese) and a peptide or protein substrate (Table 2). One must have a method to detect the conversion of substrate by detecting either the formation of phosphopeptide, phosphoprotein, the disappearance of ATP, or the formation of ADP. There are many commercially available kits and many published references describing these methodologies (Table 1). The subject of the choice of assay technologies is vast, changing and interestingly controversial (1-3) since many technologies are marketed as kits which come with strong pressure to establish “market share.” Also, in the past 15 years there has been a steady development and refinement of new kinase technologies. Examples of highlighted features in assay kits can include higher dynamic range with respect to ATP, greater sensitivity to known inhibitors, flexibility of substrate choice, statistical robustness, lowered susceptibility to artifacts and simpler assay protocol. Cost is often a consideration when choosing a technology or kit as well as the availability of instrumentation. This chapter will attempt to concisely review some of the key technologies below, highlighting the theory behind the assay, some of the underlying principles with strengths and weaknesses, in hope that the reader can make more informed decisions when reviewing the options.

Figure
Table 2. Assay Optimization Cycle and Typical Test Parameters
Radioactive Assay Technologies
Traditional kinase assays measure the transfer of the 32P from the γ position of ATP to a peptide or protein substrate results in a 32P labeled peptide or protein that may be separated away from the ATP by capture on a filter and subsequent washing. The quantity of phosphoprotein is quantified by scintillation counting (4).
The availability of [33P]ATP as an alternative to 32P provides benefits of safety and longer half-life. The lowered energy is also better suited for scintillation proximity assays (SPAs, www.perkinelmer.com; 5). SPA was a major step forward because it eliminated the need for wash steps by capturing the [33P]-labeled peptide on a functionalized scintillating crystal, usually via a biotin-streptavidin interaction.
The specific signal in the SPA is a consequence of a radiolabeled peptide or protein substrate becoming closely bound to the scintillation material. As a result, photons are given off due to a transfer of energy from the decaying 33P particle to the scintillation material. Non-specific signals (non-proximity affects) can result from decay particles emitted from free [33P]ATP molecules interacting with the scintillation material at greater distances(Figure 1).
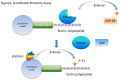
Figure 1.
Scintillation Proximity Assay
All SPAs are based upon the phenomenon of scintillation. Scintillation is an energy-transfer that results from the interaction between particles of ionizing radiation and the de-localized electrons found in conjugated aromatic hydrocarbons or in inorganic crystals. When the decay particle collides with the scintillation material, electrons are transiently elevated to higher energy levels. Because of the return to the ground state, photons are emitted. Frequently, scintillation materials are doped with fluorophores, which capture these photons (usually in the ultraviolet spectrum) and fluoresce at a “red-shifted” wavelength more “tuned” to the peak sensitivity of the detector.
Conventionally, the scintillation materials used in bioassays were liquids composed of aromatic hydrocarbons. These bioassays required a wash step before the addition of the scintillation liquid and counting in a liquid scintillation counter. With SPA technology crystals of polyvinyltoluene (PVT), Yttrium silicate (YS), polystyrene (PS) and Yttrium oxide (YOx) are used as the scintillant. These materials are functionalized with affinity tags to detect the decay particles directly in the bioassay without wash steps. The newer generation of “red-shifted” FlashPlates and SPA beads yields emission frequencies of around 615 nM, and thus, can be detected by CCD (charge-coupled device) imagers rather than photomultiplier tube (PMT) readers. The advantages of these “imaging” beads and plates lie in both the ability to simultaneously read all wells in a microtiter plate (MTP) and in the reduction of interference from colored compounds (www.perkinelmer.com).
Because of the cost of disposing of radioactive reagents and the requirement for special safety infrastructure, the use of this approach is becoming less frequent, although it presents some distinct advantages (5). First, one needs no phosphopeptide or phosphotyrosine-specific antibodies (an advantage shared with “coupled assays”, mentioned below), as the ATPγ33P is the only co-factor required. Second, because analyte detection is performed at only one emission wavelength, there are less potential sources of interference by light-absorbing compounds versus fluorescent assays. SPA techniques are well suited toward utilizing a variety of biologically relevant substrate proteins. Universal substrates that are biotinylated, such as poly-GlutamineTyrosine (polyEY), can be used for the tyrosine kinases. Generalized substrates such as myosin basic protein or casein, or specialized peptide substrates must be used for the serine-threonine kinases. A discussion of the choice of substrates in kinase assays is presented in the assay development section below.
Fluorescence Assays
Although fluorescent assays are very useful in HTS, the classic issue with these assays is that they are susceptible to interference from compounds that either absorb light in the excitation or emission range of the assay, known as an inner filter effects (6), or that are themselves fluorescent resulting in false negatives. At typical compound screening concentrations of greater than 1 µM, these types of artifacts can become significant.
There are several approaches to minimizing this interference. One approach is to use longer wavelength fluorophores (red-shifted). This reduces compound interference since most organic medicinal compounds tend to absorb at shorter wavelengths (7). The percentage of compounds that fluoresce in the blue emission (4-methyl umbelliferone) has been estimated to be as high as 5% in typical LMW compound libraries. However, this drops to <0.1% at emission wavelengths >500 nm (8). Another approach is to use as “bright” a fluorescent label as possible. Bright fluorophores have a high efficiency of energy capture and release. This means that an absorbant or fluorescent compound will have a lowered impact on the total signal of the assay. Assays with higher photon counts will tend to be less sensitive to fluorescent artifacts from compounds as compared with assays having lower photon counts. Minimizing the test compound concentration can also minimize these artifacts and one must balance the potential of compound artifacts versus the need to find weaker inhibitors by screening at higher concentrations.
The availability of anti-phosphotyrosine antibodies, anti-phosphopeptide antibodies and antibodies to fluorescent ADP analogs enabled the performance of several homogeneous methods using fluorophores, among these time-resolved Förster resonance energy transfer (TR-FRET) and fluorescence polarization (FP).
Fluorescence Anisotropy (Polarization)
Anisotropy can be measured when a fluorescent molecule is excited with polarized light. The ratio of emission intensity in each polarization plane, parallel and perpendicular relative to the excitation polarization plane, gives a measure of anisotropy, more commonly referred to in HTS as “fluorescence polarization” or FP. This anisotropy is proportional to the Brownian rotational motion of the fluorophore. Changes in anisotropy occur when the fluorescent small molecule binds to a much larger molecule affecting its rotational velocity. Kinase assays are set up using anti-phosphopeptide antibodies and labeled phosphopeptides (Figure 2, 9) or by using a metal ion affinity material to capture labeled phosphopeptides (10). The formation of the phosphopeptide in an enzymatic reaction causes an increase in binding to an antibody or affinity resin and consequently a change in anisotropy. The advantage of FP is that it requires only one small polypeptide labeled (instead of two labeled moieties as with TR-FRET or AlphaScreen).

Figure 2.
Fluorescence Anisotropy
FP assays are known to be susceptible to artifacts (11). In principle, the assays are ratiometric and should normalize for variations in total excitation energy applied as would occur with inner filter effects (see Assay Guidance Manural chapter on Spectrophotometry), and newer generations of red-shifted fluorophores should help to eliminate interference (7). However, introducing a test compound with fluorescent or absorbent properties at greater than 5 µM with the typically nanomolar concentrations of fluorescently-labeled peptide in an FP assay can significantly skew the measurements. One way to reduce this potential issue is to simultaneously collect total fluorescence data and exclude those compounds which significantly affect the total fluorescence of the assay well.
Time-Resolved (Gated) Förster Resonance Energy Transfer (TR-FRET)
These assays are based upon the use of a Europium or Terbium chelate (a transition metal-ligand complexes displaying long-lived fluorescent properties), and labeled anti-phosphopeptide or anti-phosphotyrosine antibodies that bind to phosphorylated peptides. The antibodies are usually labeled with aromatic fluorescent tags such as Cy5, Rhodamine, or fluorescent proteins such as allophycocyanin. Allophycocyanin is a light-harvesting protein which absorbs at 650 nM and emits at 660 nm (9, 12, 13). When the anti-phosphotyrosine or anti-phosphopeptide antibodies bind to a labeled phosphorylated peptide the proximity of the antibody to the labeled peptide results in a transfer of energy (Figure 3). The energy transfer is a consequence of the emission spectrum of the metal-ligand-complex overlapping with the absorption of the labeled peptide. If the donor fluor is within 7-9 nM of the acceptor fluor then Förster resonance energy transfer can occur although the optimal distance between fluorophores is also influenced by effects of proximal fluorophores on the emission lifetime in the time-gated system (14). The action of a kinase enzyme increases the concentration of phosphopeptide over time and results in an increased signal in such an assay.

Figure 3.
Time-gated Forster resonance energy transfer
TR-FRET assays have two main advantages. The first advantage is in the “time-gated” (the term “resolved” is commonly misused from a biophysical perspective) signal detection, which means that the emission is measured 100-900 µs after the initial excitation frequency is applied, resulting in a reduction in fluorescence background from the microtiter plate, buffers, and compounds. Data is acquired by multiple flashes per read, to improve the sensitivity and reproducibility of the signal. The second advantage is that the one can measure the ratio of the emission from the acceptor molecule to the emission from the donor molecule. Because of this ratiometric calculation, variations in signal due to variations in pipetting volume can be reduced. Therefore, one generally observes less inter-well variation in TR-FRET assays versus other enzyme or biomolecular assay systems (15).
Luminescent Oxygen Channeling (AlphaScreenTM, AlphaLISATM)
AlphaScreen technology, first described in 1994 by Ullman and based on the principle of luminescent oxygen channeling, has become a useful technology for kinase assays (16). AlphaScreen is a bead-based, non-radioactive, Amplified Luminescent Proximity Homogenous Assay. In this assay, a donor and an acceptor pair of 250 nm diameter reagent-coated polystyrene microbeads are brought into proximity by a biomolecular interaction of anti-phosphotyrosine and anti-peptide antibodies immobilized to these beads (www.perkinelmer.com). Irradiation of the assay mixture with a high intensity laser at 680 nm induces the conversion of ambient oxygen to a more excited singlet state by a photosensitizer present in the donor bead. The singlet oxygen molecules can diffuse up to 200 nm and, if an acceptor bead is in proximity, can react with a thioxene derivative present in this bead generating chemiluminescence at 370 nm that further activates the fluorophores contained in the same bead. The fluorophores subsequently emit light at 520-620 nm. The donor bead generates about 60,000 singlet oxygen molecules resulting in an amplified signal. Since the signal is very long-lived, with a half-life in the one second range, the detection system can be time-gated, thus eliminating short-lived background (the AlphaScreen signal is measured with a delay between illumination and detection of 20 msec). Furthermore, the detection wavelength is of a shorter wavelength than the excitation wavelength, thus further reducing the spotential for fluorescence interference. The sensitivity of the assay derives from the very low background fluorescence. The larger diffusion distance of the singlet oxygen enables the detection of binding distance up to 200 nm, whereas TR-FRET is limited to 9 nm (15), allowing the use of much larger protein substrates. A newer version of the same principle is called AlphaLISATM (Perkin-Elmer) . While AlphaScreen uses acceptor beads with an emission from rubrene, AlphaLISA acceptor beads use Europium. Thus the emission wavelength is different between AlphaScreen beads (520-620 nm) and AlphaLISA beads (615 nm). The narrow emission spectrum of AlphaLISA should, in principle, lessen interference.
Kinase assays based on the AlphaScreen principle are similar to TR-FRET assays in that they usually require a biotinylated substrate peptide and an anti-phosphoserine or tyrosine antibody. These two reagents are “sandwiched” between biotin and protein-A functionalized acceptor and donor beads. A kinase assay shows an enzyme dependent increase in antibody binding (and thus signal) over time. In some cases, the phosphorylation of an epitope will block the antibody binding which can be used as the basis of for product detection (17, 18). Like other optical assays, AlphaScreen assays are susceptible to inner filter effects (6). Additionally, compounds which react with singlet oxygen can cause false positives. One can easily re-test AlphaScreen hits in an independent AlphaScreen assay, for example measuring the effect of the compound on biotin-streptavidin bead interactions optimized to the same level of photon counts as the primary assay. Artifact compounds would be expected to inhibit this signal and can thus be eliminated as false positives.
“Coupled” Assays
“Coupled” assays are those that require the addition to the assay of more enzymes to convert a product or substrate into a detectable signal. All coupled enzyme assays share potential artifacts from “off-target” inhibition of the enzymes used to couple the reaction to a detectable product. Thus, with all coupled assays, one must be careful that inhibitors or activator compounds are not inhibiting the coupling systems. It is usually easy to design a secondary assay system to establish that this is not the case, simply by “feeding” the coupling system with suitable substrates (in the absence of kinase enzyme) and measuring the effect of putative inhibitors on the coupling system. For protein kinases, the most common manifestation of a coupled assay uses luciferase to detect ATP formation, and yet others use proteases, ATPases and other non-disclosed commercial enzymes to produce a kinase assay kit. These are briefly described below.
Protease Sensitivity Assays
Kinase substrates can become resistant to the actions of proteases due to phosphorylation. Thus, fluorescence quench assays for proteases can be used to measure kinase activity. With kinase assays, the formation of phosphopeptide inhibits the protease action on the peptide and the signal remains quenched and, therefore, lower when the kinase is active (19). Inhibiting the kinase results in an increase in protease sensitivity and an increase in signal.
Luciferase-based Kinase Assays
Because kinases convert ATP into ADP, the activity of purified kinase enzymes can be measured in a “coupled” assay, which detects the ATP depletion over time by using the phosphorescence of luciferase and luciferin (commercially sold as “Kinase-GloTM”, www.promega.com; 13, 20). Luciferases are enzymes that produce light by utilizing the high energy bonds of ATP to convert luciferin to oxyluciferin. Oxygen is consumed in the process, as follows:
- luciferin + ATP → luciferyl adenylate + PPi
- luciferyl adenylate + O2 → oxyluciferin + AMP + light
The reaction is very energy efficient: nearly all of the energy input into the reaction is transformed into light. A reduction in ATP results in a reduction in the production of photons. This type of assay has the advantage of not needing any specialized antibodies and is applicable to all kinases. It is also easy to run the assays with high ATP concentrations as a way of selecting against ATP-competitive inhibitors (21).
A potential limitation of this assay is that it is a “signal decrease assay.” In signal decrease assays, the enzyme-mediated reaction proceeds with a decrease in signal over time. As the kinase depletes the ATP over time, less light is produced by the luciferase reaction. An inhibitor compound prevents this decrease. A 50% consumption of ATP is required to obtain a two-fold signal-to-background using the 100% inhibited reaction as a control. Therefore, these assays must be run under conditions of relatively high turnover, which has the effect of slightly weakening the apparent potency of compounds (22). Another consequence of a signal decrease assay is that it can have low sensitivity for particularly low turnover enzymes (i.e., having a small kcat). For slower enzymes, one may require a long incubation time (several hours) to reach a suitable signal. With other assay formats that measure phosphopeptide formation, (especially TR-FRET, AlphaScreen), only a 2-10% conversion is required before the assay can be read, since the detecting reagents are product-specific.
Kinase assays that are coupled to luciferase have the disadvantage of being sensitive to luciferase inhibitors, which bind to the ATP-binding site of luciferase, resulting in false negatives. A kinase inhibitor which also inhibits the luciferase would result in an artificially low light signal. However, one can overcome this by additionally using a luciferase-based assay that measures ADP formation (commercially available from Promega as “ADP-Glo®”. By running Kinase-GloTM and ADP-Glo® as orthogonal assay one can identify luciferase inhibitors as these will make the signal go down in both assays while with a true kinase inhibitor the signal would go up in an Kinase-Glo assay and down in an ADP-Glo® assay (23).
The ADP-Glo system detects the ADP formed in a kinase reaction by first adding an undisclosed commercial reagent (presumably an enzyme) to deplete the ATP in the kinase reaction and then adding another undisclosed reagent to convert the ADP back into the ATP (presumably another enzyme). Coupled systems that detect ADP gain the advantage of being able to detect product formation, rather than substrate depletion, which means that small catalytic turnover percentages can be detected.
Fluorescent Coupling
Another manifestation of coupled ADP detection is the ADP-QuestTM kit (DiscoverRx, 24). In this kit, ADP is measured by a coupled enzyme reaction with pyruvate kinase and pyruvate oxidase to convert ADP to hydrogen peroxide, which is detected using a fluorogenic substrate and horseradish peroxidase. The detection wavelength is around 590 nM, which tends to be less susceptible to inner filter effects (6).
Assay Development
Much of the cited literature in this chapter offers good protocols as a basis for kinase assay development. A general strategy for assay development should include the following steps:
- 1.
Choose an appropriate readout technology.
- 2.
Synthesize, purify and characterize enzymes, substrates. Generally, highly pure preparations of kinase (greater than 98% by silver-stained SDS-PAGE or by Mass-Spectrometry) are required. It is a possibility that small amounts of contaminating kinases can result in a false detection of activity from the kinase of interest. Well-characterized preparations of enzyme are critical.
- 3.
Design a starting protocol based on prior literature or experimental information on substrate specificity. Test the protocol for enzymatic activity and use reference inhibitors when possible.
- 4.
Gain knowledge of kinetic and thermodynamic parameters as guidelines for assay optimization (25).
- 5.
Set-up “matrix” experiments varying pH, ionic strength, buffers and various other parameters mentioned below to optimize signal to background ratios (Table 2.)
- 6.
Choose volumes appropriate for HTS workstations or automated systems.
- 7.
Test a screening protocol in a pilot study and determine assay quality based on the coefficient of variations across the assay wells, the Z’ measurements (26) and dynamic range towards various control or reference compounds.
Considerations of Mechanism
Mechanism of Action of Kinase Inhibitors
When designing protein kinase assays it is important first to consider the desired mechanism of action (MOA) of the inhibitor MOA is quite a complex topic which is nevertheless well described in terms of drug discovery strategies by David Swinney (27); examples are given of drugs that work by a variety of mechanisms and fall into three basic categories: 1) competition with the substrate or ligand in an equilibrium or steady-state, 2) inhibiting at a site distinct from substrate binding or 3) inhibiting in a non-mass action equilibrium. (See Assay Guidance Manual section on mechanism of action).
A good overview of the mechanism of action of protein kinase inhibitors has been presented by Vogel and Zhong (28). Protein tyrosine kinase inhibitors can act by binding directly in the ATP binding site competitively, (type I inhibitors), but these tend to be less specific because of the shared characteristics of the ATP binding pocket among various kinases. More specificity can be attained with the type II inhibitors, which can extend into an allosteric site next to the ATP pocket and which is only available in the inactive (non-phosphorylated form) of the enzymes. Imatinib is an example of this, with a 200-fold increased potency to the inactive form of the enzyme, observed in cell based assays versus enzyme assays. Often, these inhibitors bind with a slower off-rate and on-rate due to a requirement for conformational changes.
Type III inhibitors bind to sites distal to the ATP binding site and are often inactive in simple kinase enzyme assays. This apparent inactivity is because these compounds can bind to the kinase and render it a poor substrate for an activating upstream kinase, thus disabling its activation. As a consequence, a “cascade assay” or a cell-based assay might be required.
Applying Mechanistic Principles to Assay Design
Important questions related to the desired inhibitor or agonist mechanism such as whether, in the HTS, one desires to find allosteric, competitive, slow-binding inhibitors, or inhibitors of an active or inactive form of the enzyme should be considered when designing HTS assays. These mechanisms might suggest the appropriate incubation times, substrate design and concentrations, order of addition and the appropriate recombinant construct to use. Unfortunately, the decisions of assay set up are not always straightforward. There are advantages to each type of inhibitor and it also depends largely upon the biology of the disease that is going to be treated. For example, purely ATP site competitive inhibitors might have advantages with respect to drug resistance, but disadvantages with respect to selectivity, potency and cellular activity. Small molecule inhibitors that compete with the peptide binding site are generally difficult to find since the peptide-enzyme interaction presents a large surface area. Allosteric site inhibitors might have an advantage in potency and selectivity, but also may be resistance-prone. Layered on top of this complexity is the difficulty in expressing a well-defined form of the enzyme target that is physiologically relevant.
One important consideration for assay design is that a pre-incubation step of compound with the target enzyme can be included before starting the reaction. This pre-incubation can help to favor the identification of slow, tight binding inhibitors (29), depending on the stage of the enzyme progress reaction at which the assay is stopped. The longer the reaction progress time the lower the effect of the pre-incubation step on the detection of slow-binding inhibitors. This step may add significant time to a screening protocol (pre-incubation of 15-30 minutes) and also may be difficult if the enzyme preparation is unstable. It is sometimes necessary to pre-incubate the kinase enzyme with ATP to activate it through autophosphorylation. This can be accomplished in the “stock” solution of enzyme, at stoichiometric amounts of ATP, before diluting the enzyme (and thus the ATP) into the assay. With protein kinase hit lists, it is possible to distinguish the ATP-competitive from the non-competitive inhibitors by re-screening under high and low ATP concentrations, and looking for shifts in the IC50 or percent inhibition. An ATP-dependent shift in compound potency suggests competition with ATP site according to the equation, (30)
When working with purified enzymes, it can be useful to perform a close examination of their phosphorylation state and molecular masses. Mass spectrometry is often useful for this purpose. Post-translational modifications or sequence truncations can potentially alter the compound binding sites available and can also change the structure of potential inhibitory sites. For example, with protein kinases, phosphorylations distal from the ATP binding site can inactivate the kinase whereas phosphorylations near the ATP binding site can activate the catalytic activity. Often, practice does not permit control of such situations because the purified systems are often mixtures and cannot be controlled in the commonly used recombinant expression technologies.
To favor the identification of uncompetitive or non-competitive inhibitors, one should run the assays with concentrations of substrates at least 10-fold higher than Km. To favor competitive inhibitors, one should run the assay at or below the Km values for the substrates. A balanced condition which provides for detection of all mechanisms is to keep the [S] = Km (31). Thus, for making suitable conclusions for assay design, knowledge of the kinetic and binding constants of receptors and enzymes, such as Kd, kcat ,Km, Bmax , is useful. Stoichiometric information, such as the number of enzyme molecules per assay, is also very useful since these can be used as guidelines to ensure that the assays are maximally sensitive to compounds with the desired mechanism of action. Problems in assay development often occur when the conditions required for sensitivity to the desired mechanism of action do not yield the best conditions for statistical reproducibility; therefore, compromises and balances between these two opposing factors must be often made.
The percent substrate consumption at which time the data is collected is also of importance in enzyme assay design. Typically, enzymologists like to ensure the steady-state conditions are maintained in the study of inhibitor constants such as Ki or IC50. However, many assay technologies, combined with the requirements for a robust signal to background giving a good Z′ factor (26), necessitate assay set-up where more turnover is required. This typically causes a trend toward the reduction of compound potency depending on the mechanism of action. Therefore, one must balance the need to have the most sensitive assay toward inhibition by low MW compounds with the need for sufficient signal-to-noise and signal-to-background to have statistically relevant results, for example with a coefficient of variation less than 10% around the positive and negative controls, a 5-fold difference between the positive control signal and negative control signal and/or Z’ greater than 0.5. These types of effects have been modeled by Wu and Yuan (22) and additionally confirmed by empirical determination. These investigators have found that 50% inhibition at low conversion (near zero) can translate into 31% inhibition if the assay is run at 80% conversion. IC50 values can shift as much as three fold.
Assay Optimization
It is very difficult to give specific guidelines for assay optimization in HTS due to the complexity of variables involved and because one often must balance cost, speed, sensitivity, statistical robustness, automation requirements and desired mechanism. The assay development and optimization process can be thought of as a cycle in which several variables can be tested and the parameters that give a better “reading window” can be fixed, after which further parameters can be tested under the prior fixed conditions (see Table 2). Often, one observes interactions between the various parameters. For example, the optimal detergent concentration to use may not be the same for every pH and that is why the same fixed parameters must sometimes be retested under any newly-identified optimal parameters. Furthermore, the type of optimization experiments depends upon the particular technology being used. A very important aspect of optimization is to improve the reproducibility and statistical performance of the assay, by finding conditions that increase the signal-to-noise ratio with respect to positive and negative controls, and to decrease the inter-well variations that occur due to such factors as pipetting errors and temperature gradients across the microtiter plate. In general, coefficient of variations of less than 10% and Z values greater than 0.5 are desirable.
In its simplest form, building an assay is a matter of adding several reagent solutions to a microtiter plate (MTP) with a multi-channel pipette, with various incubation times, stopping the reaction if required, and reading the MTP in a plate reader. A typical procedure might involve the following steps: 1) adding the enzyme solution to a compound-containing microtiter plate and incubating for 15 minutes; 2) adding substrates and incubating for 15 minutes to 1 hour; 3) adding a stopping reagent such as EDTA; 4) adding sensor or detector reagents, such as labeled antibodies or coupling enzymes; 5) measuring in a plate reader. The particular detector reagents to use, the assay reagent volumes, the concentrations of reagents, the incubation times, the buffer conditions (Table 2), the MTP types and the assay stopping reagents are all important parameters which need to be tested in order to obtain the very high level of reproducibility yet maintain the physiological and thermodynamic conditions to find a lead compound with the desired mechanism of action.
A very important aspect of assay development is making an appropriate choice of substrates. When possible, one should consider using a relatively physiological substrate. This means for example, using the substrate protein involved in the pathway of interest for serine-threonine kinases, rather than commonly used general substrates like maltose binding protein or, for example using a true-substrate-derived peptide sequence representing the phosphotyrosine site within the amino acid context that is found in the native substrate primary sequence, rather than an artificial substrate such as poly-Glu-Tyr. Once again this is not always practical because the natural substrates are either not well-characterized, and the artificial substrates often give a much higher turnover (kcat) and thus can yield a much more robust assay signal. If natural substrates are not readily available or robust, secondary assays can be designed using the natural substrates, to ensure the screening hits work equivalently under physiologic conditions. Additionally, it is often possible to perform a substrate screen where many random peptide sequences are tested to identify good substrates.
The danger of using solely statistical parameters (such as Z’) in assay optimization is that these do not take into account the desired physiological or biochemical mechanism of action in determining the optimal reagent concentration. For example, the optimal substrate concentration to use in an enzyme assay may not necessarily be the one that gives the best statistics with respect to reproducibility. The optimal salt concentration required for reproducibility may be far different from physiological conditions. Therefore, one must be careful to use statistical performance in a way that is consistent with the desired mechanism of action of the lead compound.
Enzyme Preparations
One cannot over-emphasize the importance of the enzyme preparation in the ability to develop a kinase assay. First, purity is important because even the slightest contamination with another enzyme can lead one to screen with a measurement of the wrong activity. Specific reference inhibitors can be used to establish that the observed catalytic activity is the correct one. Of course, when working with novel targets, such reference inhibitors may not exist. With recombinant expression systems one can generate catalytically inactive mutations to establish that the host cell is not the source of a contaminating phosphatase or kinase. In the end, however, the key requirement is to have an extremely pure and highly active enzyme preparation. If most of the enzyme molecules are inactive or if the enzyme molecules have a very low catalytic efficiency (kcat/Km), then one will need to have a high concentration of enzyme in the assay to obtain a good signal; this can limit the ability to distinguish between weaker and stronger inhibitors. If one needs to have a 100 nM enzyme concentration in the reaction, then inhibitors with a Ki of 10 nM cannot be distinguished from those with a Ki of 50 nM in a steady-state IC50 experiment.
Assay Volumes
Assay volumes usually range from 3 µL (for 1536-well MTPs) to 50 µL (384-well MTPs). Within a given total assay volume, smaller volumes of reagents are added. Frequently it is convenient to add reagents into the assay in equivalent volumes of assay buffer. As an example, for a 15 µL assay, one might add 5 µL of compound solution, 5 µL of enzyme solution, and 5 µL of substrate mix, followed by 10 µL of quench solution in a “stop” buffer. For kinase assays, this can be EDTA, which works by chelating magnesium, an essential cofactor for protein kinase catalysis. The advantages of using equal volumes are that it keeps the volumes at a level that is best for the particular pipette during automation; it minimizes the requirement for various specialized instruments and helps in the mixing of the reagents. The disadvantage of this method is that it introduces transient changes in the final concentration of reagents during the times between the various additions. In addition, enzymes are not always stable in large batches of dilute buffer required for HTS. Therefore, it can be preferred to add a low volume of a more concentrated stock solution (10X - 50X) into a higher volume of assay buffer in the well. This step often requires a liquid dispenser able to handle very low volumes of sub-microliter liquid.
Assay miniaturization helps to reduce the consumption of assay reagents, which can be very expensive. The problems encountered as one attempts to miniaturize an assay are in the change in surface to volume ratio, the lowered sensitivity and in the low volume dispensing of materials. For instance, as one moves to smaller volumes, the surfaces available for nonspecific binding increase relative to the volume. Furthermore, the smaller the volume, the less the amount of product-sensing material can be added; thus the sensitivity of the assay is reduced.
Plate Types
Generally, white plates are preferred for phosphorescent assays, such as those employing luciferase because they reflect emitted photons. Black plates are preferred for fluorescent assays because they reduce reflection. Transparent plates are generally used for imaging assays. Polystyrene is the material of choice because it can be molded reproducibly such that the plates are consistently-sized to fit into automated systems. Polystyrene tends to have some level of non-specific affinity for biomolecular reagents and, therefore, some manufacturers produce proprietary low binding plates. Polypropylene plates have low non-specific binding levels, but are more difficult to shape consistently. However, these plates are often useful as “source plates” for reagents due to their tolerance to freezing and their low level of “stickiness.” The use of 96-, low volume 96-, 384-, low volume 384-, or 1536-well depends on the assay volume and throughput that one needs. For 96-well plates, volumes range from 80-100 μl; for 384-well plates, volumes range from 15 to 100 μl; for 384-well, small-volume plates, volumes range from 4 to 10 μl; and for 1536-well MTPs, volumes range from 3 to 8 μl.
Incubation Times
Incubation times at different steps depend upon the binding kinetics or enzyme kinetics and can range anywhere from 10 minutes to 15 hours. It is generally preferred to read the reaction at a point before the reactants are depleted and the enzyme progress curve is slowing. Adding a short pre-incubation step of compound with enzyme before the initiation of the reaction allows for detection of slow-binding inhibitors, which require a conformational change of the enzyme to form a tight complex. Longer incubation steps can add significant amounts of plate processing time in HTS automation because incubation time often represents the rate-limiting step in the HTS process. Reaction rates can be increased by increasing the enzyme concentration or temperature and, in this fashion, incubation times can be reduced.
Buffers and Solvents
The concentrations of detergents, buffers, carrier proteins, reducing agents, and divalent cations can affect the specific signal and the apparent potency of compounds in concentration response curves (32). In principle, it is good to stay as close to physiological conditions as possible, but for practical reasons, there are many exceptions. For example, full-length fully regulated kinases are often impossible to express, and thus, for practical purposes non-regulated kinase domains are used in screening. Furthermore, the true physiological conditions in the microcellular environment are not always known. Sometimes carrier proteins are required for enzyme stability and to reduce nonspecific binding to reaction vessels. It is often necessary to provide additives such as protease inhibitors to prevent digestion of the assay components or phosphatase inhibitors such as vanadate to keep the product of a kinase assay intact and protected from contaminating phosphatases. Also, it is often necessary to quench assays or to add a stop reagent such as EDTA at the end of the reaction and before reading in a plate reader. A quench step or stop step is especially important when the automated HTS system does not allow for precise timing or scheduling of the assay protocol. A very good summary of solvent and buffer conditions used in kinase assay optimization is described by von Ahsen and Bomer (33).
The presence of “promiscuous” inhibitors or “aggregating” compounds in a chemical library has become a recent area of research. These compounds can cause false positives that can be reduced with certain detergents and can frequently be recognized by steep concentration response curves or by enzyme concentration-dependent shifts in the IC50 (34, 35). The exact mechanism of these false positives is not known but it is possible that these compounds induce the formation of compound-enzyme clusters, which reduce enzyme activity. Thus, it is important to have some non-denaturing detergent present in enzyme assays and to re-test hits in orthogonal assays to reduce the possibility of identifying promiscuous inhibitors in the HTS.
DMSO Concentration
In most cases, the test compounds are dissolved in DMSO and are added from a source plate into the assay. Thus, the tolerance of the assay for DMSO should be tested, by looking at the activity at various increasing concentrations of DMSO. Generally, enzymatic or biomolecular binding assays are more tolerant of high DMSO concentrations (often up to 5 - 10% DMSO). Cell based assays usually can tolerate up to 0.5% DMSO.
Detector Reagent Concentrations
Given the very large variety of assay detection methods, it is difficult to cover all the parameters needed to optimize for each detection system; however, it is important to mention that these various systems discussed in the assays readout technology section above all need to be optimized. For example, when using an antibody pair in a TR-FRET assay, it is important to find a concentration of antibodies that can trap the product efficiently at the desired time point in the reaction. In SPA, for example, an optimal SPA bead concentration should be determined empirically. In principle, the detector reagents should be present in high enough concentration to capture the analyte stoichiometrically. Having too high a concentration of detectors can be wasteful and expensive and can create higher background signals and having too low concentrations of detector reagents can compromise the dynamic range and sensitivity of the assay. A control product can sometimes be useful as a calibration standard for these types of optimization experiments. Often times, the commercial assay kit providers provide excellent protocols in optimizing the use of the detector reagents.
Pilot Screens
The final step in the assay development process is to run a “pilot” screen, where a small subset of libraries are screened to observe the hit rate, the distribution of the high signal and the low signal, typically employing the Z and Z-prime principle of Zhang and Chung (26) to assess quality. The Z factors combine the principle of signal to background ratio, coefficient of variation of the background and the coefficient of variation of the high signal into a single parameter, which gives one a general idea of the screening quality. One should be careful to closely examine the raw data and data trends from screening rather than to rely only on the Z-factor for quality control.
For pilot studies, the microtiter plates should be arrayed with reference compounds at various concentrations, to control that the screening procedure gives adequate dynamic range and sensitivity to inhibitors or activators. In general, coefficient of variations of less than 10% and Z values greater than 0.5 are desirable. The pilot studies may be used to adjust the compound concentration to obtain a reasonable hit rate; for example, one hit for every two thousand compounds tested. The hit rate will depend on the particular library screened; biased libraries may have higher hit rates than random libraries.
Acknowledgements
The author would like to acknowledge Dr. Douglas Auld for his valuable insights and comments on the draft, and Dr’s Andrew Napper, Jeff Weidner and Sittampalam Gurusingham for critical review and editing.
References
- 1.
- Lowery RG, Vogel K, Till J. ADP detection technologies. Assay Drug Dev Technol. 2010 2010 Aug;8(4):1–2. [PubMed: 20804418]
- 2.
- Goueli S., Zegzouti H., Vidugiriene J. Response to the Letter of Lowery et al. ADP Detection Technologies. Assay Drug Dev.Technol. 2010;8(4):1. [PubMed: 20804418]
- 3.
- Inglese J, Napper A, Auld D. Improving success by balanced critical evaluations of assay methods. Assay Drug Dev Technol. 2010 Aug;8(4):1. [PubMed: 20804419]
- 4.
- Gopalakrishna R, Chen ZH, Gundimeda U, Wilson JC, Anderson WB. Rapid filtration assays for protein kinase C activity and phorbol ester binding using multiwell plates with fitted filtration discs. Anal BiochemOct. 1992;206(1):24–35. [PubMed: 1456438]
- 5.
- Glickman J. F., Ferrand S. Scintillation Proximity Assays In High Throughput Screening. Assay Drug Dev.Technol. 2008:6. [PubMed: 18593378]
- 6.
- Palmier MO, Van Doren SR. Rapid determination of enzyme kinetics from fluorescence: overcoming the inner filter effect. (2007). Anal Biochem. 2007 Dec 1;371(1):43–51. Epub 2007 Jul 18. [PMC free article: PMC2211277] [PubMed: 17706587]
- 7.
- Vedvik K. L., Eliason H. C., Hoffman R. L., Gibson J. R., Kupcho K. R., Somberg R. L., Vogel K. W. Overcoming compound interference in fluorescence polarization-based kinase assays using far-red tracers. Assay.Drug Dev.Technol. 2004;2(no. 2):193–203. [PubMed: 15165515]
- 8.
- Simeonov A, Jadhav A, Thomas CJ, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. [PubMed: 18363325]
- 9.
- Newman M., Josiah S. Utilization of fluorescence polarization and time resolved fluorescence resonance energy transfer assay formats for SAR studies: Src kinase as a model system. J.Biomol.Screen. 2004;9(no. 6):525–532. [PubMed: 15452339]
- 10.
- Turek-Etienne T. C., Kober T. P., Stafford J. M., Bryant R. W. Development of a fluorescence polarization AKT serine/threonine kinase assay using an immobilized metal ion affinity-based technology. Assay.Drug Dev.Technol. 2003;1(no. 4):545–553. [PubMed: 15090251]
- 11.
- Turek-Etienne Tammy C., Small Eliza C., Soh Sharon C., Xin Tianpei A., Gaitonde Priti V., Barrabee Ellen B., Hart Richard F., Bryant Robert W. Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J.Biomol.Screening. 2003;8(no. 2):176–184. [PubMed: 12844438]
- 12.
- Moshinsky D. J., Ruslim L., Blake R. A., Tang F. A widely applicable, high-throughput TR-FRET assay for the measurement of kinase autophosphorylation: VEGFR-2 as a prototype. J.Biomol.Screen. 2003;8(no. 4):447–452. [PubMed: 14567797]
- 13.
- Schroter T., Minond D., Weiser A., Dao C., Habel J., Spicer T., Chase P., Baillargeon P., Scampavia L., Schurer S., Chung C., Mader C., Southern M., Tsinoremas N., LoGrasso P., Hodder P. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase high-throughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J.Biomol.Screen. 2008;13(no. 1):17–28. [PubMed: 18227223]
- 14.
- Vogel K. W., Vedvik K. L. Improving lanthanide-based resonance energy transfer detection by increasing donor-acceptor distances. J.Biomol.Screen. 2006;11(no. 4):439–443. [PubMed: 16751339]
- 15.
- Glickman J. F., Wu X., Mercuri R., Illy C., Bowen B. R., He Y., Sills M. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J.Biomol.Screen. 2002;7(no. 1):3–10. [PubMed: 11897050]
- 16.
- Ullman E. F., Kirakossian H., Singh S., Wu Z. P., Irvin B. R., Pease J. S., Switchenko A. C., Irvine J. D., Dafforn A., Skold C. N. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc.Natl.Acad.Sci.U.S.A. 1994;91(no. 12):5426–5430. and. [PMC free article: PMC44008] [PubMed: 8202502]
- 17.
- Von Leoprechting A., Kumpf R., Menzel S., Reulle D., Griebel R., Valler M. J., Buttner F. H. Miniaturization and validation of a high-throughput serine kinase assay using the AlphaScreen platform. J.Biomol.Screen. 2004;9(no. 8):719–725. [PubMed: 15634799]
- 18.
- Warner G., Illy C., Pedro L., Roby P., Bosse R. AlphaScreen kinase HTS platforms. Curr.Med.Chem. 2004;11(no. 6):721–730. [PubMed: 15032726]
- 19.
- Rodems, S. M., B. D. Hamman, C. Lin, J. Zhao, S. Shah, D. Heidary, L. Makings, J. H. Stack, and B. A. Pollok. 2002. A FRET-based assay platform for ultra-high density drug screening of protein kinases and phosphatases. Assay.Drug Dev.Technol. 1, no. 1 Pt 1:9-19. [PubMed: 15090152]
- 20.
- Koresawa M., Okabe T. High-throughput screening with quantitation of ATP consumption: a universal non-radioisotope, homogeneous assay for protein kinase. Assay.Drug Dev.Technol. 2004;2(no. 2):153–160. [PubMed: 15165511]
- 21.
- Kashem M. A., Nelson R. M., Yingling J. D., Pullen S. S., Prokopowicz A. S. III, Jones J. W., Wolak J. P., Rogers G. R., Morelock M. M., Snow R. J., Homon C. A., Jakes S. Three mechanistically distinct kinase assays compared: Measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors. J.Biomol.Screen. 2007;12(no. 1):70–83. [PubMed: 17166826]
- 22.
- Wu Ge, Yuan Yue, Nicholas Hodge C. Determining appropriate substrate conversion for enzymatic assays in high-throughput screening. J.Biomol.Screening. 2003;8(no. 6):694–700. [PubMed: 14711395]
- 23.
- Tanega C, Shen M, Mott BT, et al. Comparison of bioluminescent kinase assays using substrate depletion and product formation. Assay Drug Dev Technol. 2009;7:606–614. [PMC free article: PMC3096547] [PubMed: 20059377]
- 24.
- Charter NW, Kauffman L, Singh R, Eglen RM. A Generic, Homogenous Method for Measuring Kinase and Inhibitor Activity via Adenosine 5'-Diphosphate Accumulation. J. Biomol Screen. 2006;11(4):390–399. [PubMed: 16751335]
- 25.
- Pedro L, Padrós J, Beaudet L, Schubert HD, Gillardon F, Dahan S. Development of a high-throughput AlphaScreen assay measuring full-length LRRK2(G2019S) kinase activity using moesin protein substrate. Anal Biochem. 2010 Sep 1;404(1):45–51. Epub 2010 Apr 29. [PubMed: 20434426]
- 26.
- Zhang J. H., Chung T. D., Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J.Biomol.Screen. 1999;4(no. 2):67–73. [PubMed: 10838414]
- 27.
- Swinney D. C. Biochemical mechanisms of drug action: what does it take for success? Nat.Rev.Drug Discov. 2004;3(no. 9):801–808. [PubMed: 15340390]
- 28.
- Vogel Kurt W., Zhong Zhong, Bi Kun, Pollok Brian A. Developing assays for kinase drug discovery - where have the advances come from? Expert Opin.Drug Discovery. 2008;3(no. 1):115–129. [PubMed: 23480143]
- 29.
- Glickman J. F., Schmid A. Farnesyl pyrophosphate synthase: real-time kinetics and inhibition by nitrogen-containing bisphosphonates in a scintillation assay. Assay.Drug Dev.Technol. 2007;5(no. 2):205–214. [PubMed: 17477829]
- 30.
- Segal, I, 1975. Enzyme Kinetics: Behavior and analysis of rapid equilibrium and steady-state Enzyme systems. New York, Wiley and Sons, p106.
- 31.
- Yang J, Copeland RA, Lai Z. Defining balanced conditions for inhibitor screening assays that target bisubstrate enzymes. J Biomol Screen. 2009;14:111–120. [PubMed: 19196704]
- 32.
- Schröter A, Tränkle C, Mohr K. Modes of allosteric interactions with free and [3H]N-methylscopolamine-occupied muscarinic M2 receptors as deduced from buffer-dependent potency shifts. Naunyn Schmiedebergs Arch Pharmacol. 2000 Dec;362(6):512–9. [PubMed: 11138843]
- 33.
- Von Ahsen Oliver, Boemer Ulf. High-throughput screening for kinase inhibitors. ChemBioChem. 2005;6(no. 3):481–490. [PubMed: 15742384]
- 34.
- Feng B. Y., Shoichet B. K. A detergent-based assay for the detection of promiscuous inhibitors. Nat.Protoc. 2006;1(no. 2):550–553. [PMC free article: PMC1544377] [PubMed: 17191086]
- 35.
- Shoichet B. K. Interpreting steep dose-response curves in early inhibitor discovery. J.Med.Chem. 2006;49(no. 25):7274–7277. [PubMed: 17149857]
Additional References:
- Daub H., Specht K., Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat.Rev.Drug Discov. 2004;3(no. 12):1001–1010. [PubMed: 15573099]
- Johnston P. A., Foster C. A., Shun T. Y., Skoko J. J., Shinde S., Wipf P., Lazo J. S. Development and implementation of a 384-well homogeneous fluorescence intensity high-throughput screening assay to identify mitogen-activated protein kinase phosphatase-1 dual-specificity protein phosphatase inhibitors. Assay.Drug Dev.Technol. 2007;5(no. 3):319–332. [PubMed: 17638532]
- Lakowicz, J. R. 2006. Principles of Fluorescence Spectroscopy. 3rd ed. Berlin: Springer.
- Lawrence David S., Wang Qunzhao. Seeing is believing: peptide-based fluorescent sensors of protein tyrosine kinase activity. ChemBioChem. 2007;8(no. 4):373–378. [PMC free article: PMC3057110] [PubMed: 17243187]
- Olive D. M. Quantitative methods for the analysis of protein phosphorylation in drug development. Expert.Rev.Proteomics. 2004;1(no. 3):327–341. [PubMed: 15966829]
- Parker Gregory J., Law Tong Lin, Lenoch Francis J., Bolger Randall E. Development of high throughput screening assays using fluorescence polarization: nuclear receptor-ligand-binding and kinase/phosphatase assays. J.Biomol.Screening. 2000;5(no. 2):77–88. [PubMed: 10803607]
- Zegzouti H, Zdanovskaia M, Hsiao K, Goueli SA.2009. ADP-Glo: A Bioluminescent and homogeneous ADP monitoring assay for kinases. Assay Drug Dev Technol. Dec;7(6):560-72. PubMed PMID: 20105026. [PubMed: 20105026]
- Review Basics of Enzymatic Assays for HTS.[Assay Guidance Manual. 2004]Review Basics of Enzymatic Assays for HTS.Brooks HB, Geeganage S, Kahl SD, Montrose C, Sittampalam S, Smith MC, Weidner JR. Assay Guidance Manual. 2004
- Hypertonic saline activates protein tyrosine kinases and mitogen-activated protein kinase p38 in T-cells.[J Trauma. 1997]Hypertonic saline activates protein tyrosine kinases and mitogen-activated protein kinase p38 in T-cells.Junger WG, Hoyt DB, Hamreus M, Liu FC, Herdon-Remelius C, Junger W, Altman A. J Trauma. 1997 Mar; 42(3):437-43; discussion 443-5.
- Development of a high-throughput screening method for LIM kinase 1 using a luciferase-based assay of ATP consumption.[J Biomol Screen. 2012]Development of a high-throughput screening method for LIM kinase 1 using a luciferase-based assay of ATP consumption.Mezna M, Wong AC, Ainger M, Scott RW, Hammonds T, Olson MF. J Biomol Screen. 2012 Apr; 17(4):460-8. Epub 2011 Dec 7.
- Review Development and application of PI3K assays for novel drug discovery.[Expert Opin Drug Discov. 2015]Review Development and application of PI3K assays for novel drug discovery.Yanamandra M, Mitra S, Giri A. Expert Opin Drug Discov. 2015 Feb; 10(2):171-86. Epub 2014 Dec 30.
- Screening in a cell-based assay for inhibitors of microglial nitric oxide production reveals calmodulin-regulated protein kinases as potential drug discovery targets.[Brain Res. 1999]Screening in a cell-based assay for inhibitors of microglial nitric oxide production reveals calmodulin-regulated protein kinases as potential drug discovery targets.Mirzoeva S, Koppal T, Petrova TV, Lukas TJ, Watterson DM, Van Eldik LJ. Brain Res. 1999 Oct 9; 844(1-2):126-34.
- Assay Development for Protein Kinase Enzymes - Assay Guidance ManualAssay Development for Protein Kinase Enzymes - Assay Guidance Manual
Your browsing activity is empty.
Activity recording is turned off.
See more...
