NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Institute of Medicine (US) Forum on Microbial Threats. Addressing Foodborne Threats to Health: Policies, Practices, and Global Coordination: Workshop Summary. Washington (DC): National Academies Press (US); 2006.

Addressing Foodborne Threats to Health: Policies, Practices, and Global Coordination: Workshop Summary.
Show detailsOVERVIEW
Foodborne illness is estimated to affect more than 76 million people in the United States each year, resulting in 325,000 hospitalizations and 5,200 deaths, but its true incidence is unknown (Mead et al., 1999). Because foodborne disease is difficult to diagnose, the vast majority of these illnesses and more than half of such deaths are attributed to “unknown agents” (Mead et al., 1999). The annual cost of medical expenses and productivity losses associated with the five most prevalent, diagnosable foodborne illnesses is nearly $7 billion (Vogt, 2005).
Many people with symptoms of foodborne illness do not seek medical attention, further contributing to underdiagnosis. These circumstances, in addition to the rapid distribution of food on both a national and global scale, make it nearly impossible to detect even a large foodborne outbreak in time to limit its impact; see, for example, the description of the 1994 Salmonella outbreak in ice cream, described by Osterholm in Chapter 1. Most often, outbreak investigations occur after the fact. However, as the papers in this chapter illustrate, findings from outbreak investigations enable public health authorities to identify new foodborne pathogens, trace their entry into the food chain, and thereby reveal opportunities to improve food safety.
The first contribution to this chapter, by Robert Tauxe of the Centers for Disease Control and Prevention (CDC), provides an overview of the foodborne threat spectrum and the practices of public health surveillance by which these microbes, and the burden of disease they cause, have become known. Tauxe explores several recent advancements in this field, including the development of information networks for foodborne disease surveillance (see also Besser in Chapter 5) and enhanced outbreak investigations, and their probable link to recent reductions in cases of several major foodborne diseases.
Despite these improvements, the processes of outbreak detection and investigation remain highly challenging, as illustrated in the case studies that make up the remainder of this chapter. The first two papers, by Barbara Herwaldt of the CDC and Roberta Hammond and Dean Bodager of the Florida Department of Health, describe their experiences investigating a relatively new foodborne threat: the coccidian parasite Cyclospora cayetanensis. Little was known about the organism when, in the mid-1990s, large, multistate outbreaks of gastroenteritis were recognized. Herwaldt and public health colleagues eventually traced these outbreaks to raspberries from Guatemala, where Cyclospora infection is endemic. Several other types of fresh produce have also been identified as vehicles for cyclosporiasis outbreaks. Herwaldt analyzes the challenges presented by food-borne outbreaks (in general, as well as the specific difficulties associated with C. cayetanensis) and draws important lessons for the future of public health.
In the subsequent paper, Hammond and Bodager describe the complexities of a recent C. cayetanensis investigation. Triggered by an early 2005 report from a private lab of an unusually large number of infections, the investigation ultimately involved county health departments throughout Florida, three different state agencies that regulate food in Florida, and two federal agencies: the CDC and the Food and Drug Administration (FDA). The investigators determined that imported basil provided the vehicle for the parasite; like raspberries, basil is a “stealth” ingredient that many people do not recognize or (because such foods are often served as garnishes) easily forget. Such accounts illustrate the importance of examining seemingly unrelated cases of apparent foodborne illnesses as indicators of outbreaks and pursuing them to their sources through timely and thorough investigation.
The pathogen discussed in the chapter’s final contribution, the hepatitis A virus (HAV), is far better characterized than Cyclospora, yet its investigators are faced with a similar array of challenges. This paper, by workshop speaker Beth Bell and Anthony Fiore of the CDC, describes a series of hepatitis A outbreaks in late 2003 that included the largest such outbreak reported in the United States. It involved over 600 patrons of a single Pennsylvania restaurant, and ultimately led the FDA to ban imports from the Mexican farms that grew the tainted green onions that caused the outbreak. Investigators were aided by molecular methods for HAV detection (comparable methods do not exist for Cyclospora), but Bell and Fiore note several characteristics of routine hepatitis A surveillance and of the infection itself that continue to hinder its detection and control. The authors conclude that foodborne HAV infection (and those of other enteric pathogens) may be best prevented on the farm by reducing the contamination of produce with fecal material.
Taken as a whole, the papers in this chapter demonstrate both the crucial importance and the daunting difficulty of conducting foodborne outbreak investigations. The success of such investigations depends to a large extent on public and private laboratories that must have adequate resources if they are to quickly and accurately detect threats to our food supply. Indeed, Tauxe observes that future advancements in the detection and investigation of foodborne illness are less likely to be achieved through technical innovation than through the strengthening of public health infrastructure.
THE BURDEN OF ILLNESS ASSOCIATED WITH FOODBORNE THREATS TO HEALTH, AND THE CHALLENGE OF PREVENTION
Robert V. Tauxe, M.D., M.P.H.1
Centers for Disease Control and Prevention2
Few human endeavors are more complex than the constant, daily, and varied effort to produce and prepare the foods we eat. The many cultural traditions and changing tastes introduce new foods and food-making processes to growing populations around the world. As a result, the foodborne diseases that follow the contamination of the food supply with any of a large number of microbes and toxins present similarly evolving challenges. A new foodborne disease may emerge when a previously unknown pathogen appears in a reservoir related to the food supply or when transmission through a new foodborne pathway is documented for a known pathogen. When a new foodborne disease appears, there is a natural history to the challenge, starting with first detection and description; the development of means to diagnose and treat the new infection; investigations into the sources, reservoirs, and transmission pathways; and finally prevention stratagems that improve to the point that the disease no longer presents an important problem. Each of the many known foodborne diseases is somewhere on this progression, and more are likely to be appreciated in the future. The spectrum of foodborne diseases is a dynamic range of threats.
An array of bacterial, viral, and parasitic pathogens that cause foodborne infections are currently recognized as public health problems in the United States. Among these, an important number have only been recognized as foodborne pathogens in the last three decades (Table 3-1). Some were first detected as pathogens in recent times and may represent the evolution of new combinations of virulence properties. For example, E. coli O157:H7, not detected at all before the 1970s and first recognized as a cause of human illness in 1982, became a major foodborne disease with a recognized bovine reservoir on several continents by the 1990s (Griffin and Tauxe, 1991). This pathogen evolved from precursors with far less pathogenic potential as the result of several phage-induced mutations (Wick et al., 2005). Though the timing of these modifications remains unproven, several have noted that mobilization of phages and of other transferable genetic elements could be linked to exposure to antimicrobial agents (Zhang et al., 2000; LeFebvre et al., 2005) and therefore perhaps linked to relatively recent changes in agriculture. Another recent example is the emergence of an entirely new toxigenic serotype of Vibrio cholerae with epidemic potential. This serotype, O139, first appeared in 1992 in India, and spread rapidly through much of South and Southeast Asia where it was transmitted through water and food (Hoge et al., 1996). This serotype appears to have evolved as the result of a horizontal transfer of the genes that produce the O-antigen, possibly from another Vibrio, into several strains of the dominant strain of epidemic toxigenic V. cholerae O1 (Faruque et al., 2003).
TABLE 3-1
Foodborne Pathogens in the United States.
Other pathogens were recognized as human pathogens well before they were linked to foodborne transmission. For example, Listeria monocytogenes, first described as a cause of severe invasive infections in humans in the 1930s, was first linked to foodborne transmission in 1981 in an outbreak associated with cole slaw (Schlech et al., 1983), and more recently it was documented to be primarily foodborne (Slutsker et al., 2000). Campylobacter jejuni, described as a cause of invasive infection in immunocompromised hosts in the 1950s, was shown in 1977 to be a common enteric pathogen in normal hosts; the importance of foodborne transmission was established by 1980 (Blaser et al., 1983). The more recent observations of the parasite Cyclospora cayetanensis show how a pathogen that was geographically restricted to remote and third-world locations may leap to the forefront as a new food safety challenge, as summarized elsewhere in this report. This means that the new and emerging foodborne pathogens observed elsewhere in the world are of substantial interest and may offer a view into our future. The recent reports of outbreaks of Yersinia pseudotuberculosis associated with lettuce in Finland and of hepatitis E infection associated with swine in Japan are worthy of our attention (Nuorti et al., 2004; Yazaki et al., 2003).
Still others represent the recrudescence of foodborne challenges long brought under control, as changing tastes and patterns of trade reintroduce pathogens to the public that we last saw as a significant problem many decades ago. Souvenir seafood brought back in suitcases led to foodborne cholera in New Jersey in the 1990s (Finelli et al., 1992). The recent appearance of bovine tuberculosis in New York City may be a result of the rapid shipment of homemade cheeses from Latin America, made traditionally with unpasteurized milk (CDC, 2005a). The specter of an intentional attack on the population through the food supply has added other pathogens—new and old—to the list of potential threats (Sobel et al., 2002).
We can anticipate new challenges to continue to emerge. A robust and flexible public health surveillance system is an important part of how we will detect, characterize, and ultimately prevent these new challenges.
Public Health Surveillance
Public health surveillance is conducted to define the magnitude and burden of a disease that needs public health action, to identify and investigate outbreaks so that control measures can be rapidly implemented and issues in need of further research swiftly identified, and to measure the impact of control and prevention efforts. The public health surveillance of infections that are likely to be foodborne now includes a substantial list of pathogens whose diagnosis is to be reported to public health authorities, and a new set of national networks for characterizing the pathogens and the illnesses they cause. The recent improvements in surveillance have been summarized in detail in a recent Institute of Medicine (IOM) publication (IOM/NRC, 2003). The following is a brief sketch of some of the improvements.
The primary authority for surveillance rests with the state health departments, which gather information from cities and counties and operate most of the public health laboratories. State and local notifiable diseases laws request or require clinicians and laboratories to report specific infections and to refer isolates of some pathogens to the public health laboratory for further characterization. These laws also typically require the reporting of unusual clusters or outbreaks of disease. In addition, many jurisdictions maintain complaint lines, to which concerned citizens may directly report illnesses or observations they think may need public health attention. Some food testing occurs in the course of routine inspections and as part of process monitoring within food production. This testing may also provide some information about the status of the food supply, though its purpose is usually the ongoing verification of process control, not safety testing of each lot.
Since 1996, the public health surveillance system for foodborne diseases has been strengthened in several ways. Several diseases were added to the standard notifiable disease reporting system, including non-O157 Shiga toxin-producing E. coli, hemolytic uremic syndrome, Cyclospora cayetanensis, and Listeria monocytogenes. The routine public health serotyping of Salmonella and Shigella was strengthened by the production and distribution of new antisera and training in their use; now new DNA sequence-based methods are being developed for more rapid identification of the serotype of Salmonella (McQuiston et al., 2004). Public health monitoring of antimicrobial resistance in several enteric bacterial pathogens has been implemented in parallel with monitoring of resistance in the same pathogens isolated from animals and foods, leading to the identification of such hazards as fluoroquinolone-resistant Campylobacter jejuni and multi-drug resistant strains of Salmonella enteriticas serotype Typhimurium and Salmonella enteriticas serotype Newport (Holmes and Chiller, 2004).
The reporting of outbreaks of foodborne diseases from local and state health departments has been improved by standardized and rapid reporting via the Internet and the Electronic Foodborne Outbreak Reporting System (CDC, 2005d). Enhanced surveillance, including a new collection form and improved close-out procedures doubled the number of foodborne outbreaks reported to more than 1,200 outbreaks each year (Figure 3-1). Now the Electronic Foodborne Outbreak Reporting System has changed an old and slow paper-based system into a more rapid reporting that makes it likely that a cluster of similar outbreaks occurring in several parts of the country at once will be detected and flagged, and also increasing the utility of the surveillance data to track trends in specific foodborne outbreak categories.
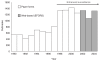
FIGURE 3-1
Reported outbreaks of foodborne diseases, 1990–2004, United States. SOURCE: Adapted from CDC (2006b).
PulseNet, CDC’s national network for subtyping foodborne bacterial pathogens, has been implemented in all 50 states and a growing number of large city health departments, as well as in the laboratories of the food regulatory agencies at the U.S. Department of Agriculture (USDA) and the FDA (Gerner-Smidt et al., 2006). This network relies on the submission of isolates of E. coli O157:H7, Listeria monocytogenes, Salmonella, and other bacterial pathogens from clinical laboratories to the public health laboratory, where the DNA “fingerprint” is determined using pulsed-field gel electrophoresis. Automated comparison of the digitized DNA pattern with the growing state and national database can swiftly identify strains (and therefore cases) that might be related, detecting clusters spread across multistate jurisdictions that might otherwise have been missed completely. In the 1960s, Salmonella serotyping transformed surveillance for that organism by increasing the signal-to-noise ratio and making it possible to pick out outbreaks of one serotype from the background noise of all salmonellosis (CDC, 1965). Now PulseNet provides an additional specificity, with a generally applicable tool for identifying clusters of infections that are likely to be related, even within a single closely-related serotype such as E. coli O157:H7, or within individual Salmonella serotypes. PulseNet test protocols have now been developed for seven bacterial foodborne pathogens, as well as for Yersinia pestis and F. tularensis.
PulseNet protocols have now been adopted in Canada, Japan, Australia and other countries and are the heart of international networks for surveillance in Europe, Asia and the Pacific, and Latin America (Swaminathan et al., 2006). This will enhance our own prevention capacity. For example, in 2004, public health laboratories in Japan detected a small cluster of E. coli O157:H7 infections in Okinawa that they linked to consuming ground beef from the commissary at a U.S. military base there, and an indistinguishable E. coli was detected in ground beef in Japan, which came from the United States (CDC, 2005b). The notification by Japan led to recall of 90,000 pounds of ground beef shipped to the military and other institutions in the United States. The same strain was also identified in two persons in the United States who did eat beef the origin of which was not traceable, and who would not otherwise have been linked.
In the future, routine usage of multilocus variable number tandem repeat assays or other sequence based-methods in state health department laboratories will further refine the speed and precision of the network. However, the promise of real-time results is more dependent on resources, rather than technology, including the vital participation of the private clinical laboratory sector to refer strains rapidly to the public health laboratory and on the laboratory support within the state health department to run the tests swiftly.
Another major advance in foodborne surveillance has been FoodNet, the active sentinel site surveillance system for foodborne illness (Allos et al., 2004). While PulseNet enhances the ability of all states to detect clusters and investigate outbreaks, FoodNet is focused on developing standard and detailed surveillance data on sporadic (nonoutbreak-associated cases) in 10 sites around the country, now representing 14 percent of the U.S. population. Though sporadic cases are far more common than those that are associated with outbreaks, they receive far less attention in general. Active surveillance means that the health department regularly contacts the clinical laboratories to collect reports of what has been diagnosed, rather than relying on the laboratories to report them. In addition FoodNet conducts specialized surveys of the clinical laboratories, of the general population, and of other groups to obtain measures of the frequency of gastroenteritis in general, of specific diagnostic tests, and other measures important to interpreting surveillance data. Data from FoodNet have been critical to refining the overall estimates of the burden of foodborne disease and to tracking trends in specific infections over time. For example, between 1996 and 2004, FoodNet documented a 42 percent decline in diagnosed E. coli O157 infections, decreasing to 0.9 per 100,000 in the year 2004; a 40 percent decline in Listeria infections; and a 31 percent decline in Campylobacter infections (CDC, 2005c). With case-control and other studies, FoodNet also defines the association between infections and specific foods, contributing to the attribution of the burden of specific infections to foods. Increasingly, FoodNet serves as a platform for developing and evaluating improved public health surveillance and investigative and prevention strategies.
Estimating the Burden of Foodborne Diseases
The health burden of an infection includes the morbidity it causes, the hospitalization and other medical care that results, and the mortality, among other measures. Estimating this burden for a given pathogen means going beyond the reported cases. To contribute a reported case, the person must become ill, must seek medical care, the physician must ask for a laboratory test, the patient must provide a specimen for diagnostic study, the specimen must yield evidence of the pathogen, and the case must be reported. Slippage at each point means that the diagnosed cases are likely to represent only a small fraction of the cases that actually occur. Other measures of severe infection, such as hospital discharge summary records and cause of death as reported on death certificates, may be used to estimate the total number of hospitalizations and deaths due to acute enteric disease, but these measures significantly underreport specific infections, as laboratory diagnoses may often not be reflected in the discharge or death certificate coding. In 1999, we published a report estimating the actual acute health burden of foodborne disease in the United States (Mead et al., 1999).
These estimates were assembled from a variety of data collected by FoodNet and other sources. FoodNet population surveys measure the number of cases of acute gastroenteritis that actually occur and the proportion of these that seek care and are cultured (Herikstad et al., 2002). The FoodNet clinical laboratory surveys measure the likelihood that a specimen will be routinely tested for say, Salmonella or Campylobacter or E. coli O157 (Voetsch et al., 2004a). This information can then be used to amplify the number of cases that are diagnosed and reported; in this way FoodNet estimated that there are actually 38 cases of salmonellosis for every one that is diagnosed and reported (Voetsch et al., 2004b). FoodNet data also provide the number of diagnosed salmonellosis cases that lead to hospitalization and the number that lead to death. Doubling that number to account for cases that were not cultured provides a conservative estimate of the total number of hospitalizations and deaths. Using similar data and assumptions, the incidence of other infections under surveillance by FoodNet can also be estimated, and by use of a uniform set of assumptions and expert opinion it is possible to estimate the overall burden of known enteric infections at some 39 million infections per year (Mead et al., 1999).
The next step was to estimate the proportion of these infections that are transmitted through food, rather than through water, direct contact with ill children, or other pathways. The estimated proportion of infections that are transmitted through foods varied by pathogen, and in sum was 38 percent. Thus, of 39 million enteric infections estimated to be caused by the known enteric pathogens, 16 million were attributed to food. A curious observation is that the estimate of acute enteric illness developed pathogen by pathogen (annual incidence of 39 million cases) is substantially less than the total amount of acute gastroenteritis in the population estimated by population survey (annual incidence of 211 million cases) (Mead et al., 1999). This “diagnostic gap” suggests that there are more pathogens yet to be discovered (Tauxe, 2002). The fraction of these other cases not accounted for by known pathogens that might be attributed to food is not directly measurable. The authors of the 1999 estimate chose 38 percent, the summary statistic for the known pathogens, as the best point estimate of what it might be for the other acute illnesses not accounted for by known pathogens. The final estimate, 76 million illnesses, 323,000 hospitalizations, and 5,000 deaths, refers to the year 1997. This comprehensive estimate is now being revised in a similar stepwise approach, starting with the measurement of the overall burden of acute gastroenteritis and with more refined and pathogen-specific approaches to the estimates of unreported illness.
There are other ways of measuring the burden of unreported illness. In the United Kingdom, the Intestinal Infectious Diseases study empanelled a group of citizens who recorded their symptoms prospectively and provided stool specimens for even mild cases of diarrheal illness (Wheeler et al., 1999). The Dutch SENSOR study followed a similar strategy, working with a group of sentinel general practitioners and their patients (de Wit et al., 2001). Both European approaches depended on the national healthcare system itself to provide a population-based framework, and both were sufficiently expensive that they have not been repeated. There are also measures of burden other than simple counts of cases, hospitalizations, and deaths. For example, the health-related costs for the principal bacterial foodborne pathogens (Salmonella, Campylobacter, E. coli O157, other Shiga-toxin-producing E. coli, and Listeria monocytogenes) have been estimated to be $6.9 billion (ERS, 2000). The cost to society associated with the estimated number of deaths that were not attributed to known etiologies could be as high as $17 billion, underlining the need for further refinement of this sector of the estimate (Frenzen, 2004). Inclusion of the postinfectious sequelae in the estimate can also greatly increase the economic burden. A detailed model developed for Campylobacter in the Netherlands included the burden of postinfectious arthritides and Guillain-Barre syndrome and measured the burden in disability-adjusted life years; this estimate indicated that a greater burden was due to the sequelae, rather than the acute illness (Havelaar et al., 2005). The industry costs of disrupted trade and development that can be occasioned by foodborne illness can be enormous, though they usually do not appear on the public health ledger. The costs of antimicrobial resistance associated with foodborne exposures have also not been estimated, but they might include the cost of illness caused by resistant foodborne pathogens and the costs related to the spread of transmissible resistance genes that are present in commensal organisms in the food supply, from which they may transfer to human pathogens.
Prevention of Emerging Foodborne Threats: The Importance of the Outbreak Investigation
The prevention of foodborne diseases in general is a complex effort, involving many different actors along the chain of production from the farm to food service. There are many different pathogens involved, almost none of which are vaccine preventable in the final consumer. Educating consumers, food handlers, and producers about their role in preventing foodborne disease is important, but not sufficient. Contamination of food can occur at many points from farm to table. Often the key to prevention is to understand those mechanisms of contamination well enough to prevent them, before the food reaches the final consumer. Investigation of contamination events, and especially investigation of outbreaks, is critical to understanding the mechanisms of contamination. Prevention often means reengineering food processes and policies for safety, usually with a focus on a specific food and/or pathogen.
The foodborne outbreak investigation is thus a major driver for enhancing overall food safety. When an outbreak is detected, the first priority is to learn enough to prevent further cases from occurring in the current outbreak. However, it is also an opportunity to learn something new, and to open research agendas with impact far beyond the one outbreak. Many foodborne pathogens were first identified in the course of an outbreak investigation. A new combination of pathogen and food may be revealed that needs further study by food scientists, animal and plant pathologists, as well as medical researchers. Just as the National Transportation Safety Board investigates a plane crash thoroughly after the fact to learn how to prevent similar ones, careful investigation even after an outbreak is over can define gaps in the system, stimulate further specific research, and indicate the needs for new processes or regulations. New combinations of specific pathogens and foods identified by outbreak investigations have been critical to guiding research and prevention (see Table 3-2).
TABLE 3-2
Some New Pathogen-Food Combinations Characterized During Outbreak Investigations in the United States.
As the surveillance systems that we use in the United States have been enhanced in the last 10 years, we have observed a change in the number and nature of outbreaks detected. This is a paradox of surveillance: making surveillance better often reveals more of the problem, so that the actual public health burden appears worse. For example, as noted above, the number of foodborne outbreaks reported through the Electronic Foodborne Outbreak Reporting System doubled following relatively simple improvements in process and participation. PulseNet has caused a more substantial change in the nature of the outbreaks detected. By increasing the signal-to-noise ratio for specific pathogen subtypes, PulseNet makes it far more likely that geographically diffuse outbreaks will be detected. Those diffuse outbreaks are particularly instructive.
PulseNet has had a profound impact on the kind of outbreaks that have been detected because the nature of the outbreaks detected depends critically on the methods used to detect them. If outbreaks only come to the attention of public health when concerned citizens, physicians, or healthcare facilities report them, then only large and locally apparent outbreaks are likely to be detected. These classic point source outbreaks often affect a single group of people in a single town or city, following a single meal, with a substantial attack rate. Investigating this outbreak proceeds with local authority, and the food-handling problems that are identified are often local in scope. Although important, these investigations may have greatest impact at the local level.
The use of molecular subtyping to compare strains across many jurisdictions has revealed an entirely different kind of outbreak in which a dispersed group of persons who do not know each other are affected at the same time with the same infecting organism in many different locations. In this scenario, no local listening post may perceive more than a few cases, and the local increase is often not apparent against the background of cases. Although each individual case may appear to be sporadic, the outbreak may in fact be very large but dispersed. Investigating these dispersed scenario outbreaks requires the coordinated efforts of many health authorities acting in concert and pooling the information. Though difficult to detect and to investigate, the findings of these outbreaks can be of particular importance. The dispersion may well reflect a contamination event high in the food’s chain of production, not just a problem in the final kitchen. Identifying that event can instruct the industry and regulatory authorities about a flaw in the system that was previously unappreciated. Correcting it can lead to lasting and generalized protection across the country.
This means that improved detection and investigation can serve to probe the safety of the food production system at several levels. These investigations, providing information about gaps in the food safety system, drive the cycle of prevention faster and reduce the overall number of infections. The results of enhanced prevention can be seen in the recent declines in the incidence of infections with Listeria monocytogenes and E. coli O157, the two pathogens tracked most intensively by PulseNet. Following the institution of PulseNet surveillance for Listeria monocytogenes, there was an important increase in the number of outbreaks detected (Figure 3-2). Many of these were related to processed meats, focusing prevention efforts on that sector; incidence declined to an all time low of 2.7 per million in 2004, a drop of 40 percent since the baseline period 1996–1998 (CDC, 2005c). The incidence of E. coli O157 infections began to decrease sharply after 2002, as the repeated investigations of pulsed-field gel electrophoresis clusters focused attention on more specific controls at the level of ground beef. By 2004, the incidence of E. coli O157 infections as measured in FoodNet had dropped 42 percent since the baseline period of 1996–1998, and was 0.9 per 100,0000, below the goals set by Healthy People 2010. It is doubtful that such progress would have been made without PulseNet.

FIGURE 3-2
Reported incidence of Listeria monocytogenes infections and reported outbreaks of listeriosis, United States, 1986–2004. SOURCES: Adapted from Tappero et al. (1995); CDC (2006a,).
The most recent outbreak surveillance information published for E. coli O157:H7 also illustrates how improved surveillance can first produce a sharp increase in reported outbreaks, followed by a drop as better prevention strategies take effect (Figure 3-3) (Rangel et al., 2005). In the 1980s, E. coli O157 outbreaks of infection were rare, perhaps because the pathogen itself was less common, but also because it was not likely to be diagnosed or reported. Washington, the first state to make it a notifiable condition, did so in 1988. After the large Western states outbreak of 1993, centered in Washington, many other states made it notifiable, and it became nationally notifiable in 1994. At the same time, an education effort targeting clinical laboratories promoted simple laboratory screening for the pathogen. It is not surprising that the number of reported outbreaks jumped to more than 30 in 1994, and then began to decline, as many in the fast-food industry and homes changed burger cooking procedures to avoid undercooking. In 1996–1997, in the first FoodNet case-control study of sporadic E. coli O157:H7 infections, eating burgers at a fast-food restaurant was no longer associated with illness, though it had been in earlier studies (Kassenborg et al., 2004). Following the launch of PulseNet, the number of reported outbreaks more than doubled again in 1998, and since then has generally trended downwards as other prevention measures have been enacted. As noted above, the biggest decrease may have happened after 2002, after new procedures to reduce the contamination of ground beef were implemented, though the impact of those on E. coli O157:H7 outbreaks has not yet been summarized in print.
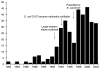
FIGURE 3-3
Reported outbreaks of E. coli O157 infections, United States, 1982–2002. SOURCE: Rangel et al. (2005).
Constraints and Limitations on Using Outbreaks to Drive Control and Prevention
Detecting and investigating foodborne outbreaks triggers public health intervention, but as a prevention system, this has built in delays. Most obviously, it is not activated until after people are exposed, become ill, and the outbreak is detected. Often the outbreak is actually over by the time it is detected, making the outbreak investigation itself a blunt instrument for achieving control over single brief exposures to contamination. There are biology-dependent delays, like the incubation period between the moment of exposure and the onset of symptoms, which can vary from less than a day for some pathogens to several weeks for others. Delays in diagnosis may depend on when the typical patient feels sufficiently ill to consult a physician and on how long the laboratory tests take to yield a diagnosis. Signal delay depends on the time it takes to accumulate enough cases in one database to be detectable as a distinct cluster and may be longer if cases are more dispersed. There are also the resource-dependent delays that depend critically on support that surveillance gets in the private and public sectors. Clinical laboratories may batch isolates for shipment to the public health laboratory to cut shipping costs, adding delays. The speed of testing strains in public health laboratories also depends on the available resources. Interviewing cases and tracing implicated foods back to their sources depends on the availability of trained and supported investigative staff, for whom this is part of their core work duties, not a distraction from the “real” work they must accomplish.
Many state and local health departments lack sufficient capacity to effectively investigate outbreaks. For example, in a recently published survey of state health departments, 88 percent reported there were barriers to more active case finding, and 30 percent reported that they lacked adequate epidemiological staff to conduct investigations. Outbreaks go uninvestigated for a number of reasons, the most common of which are delayed notification (83 percent of those respondents) and lack of staff (67 percent). Many state public health laboratories are also understaffed and under supported, making real-time testing of submitted strains difficult and leaving them with little surge capacity for emergencies (APHL, 2003).
Pooling Resources
The fundamental ability to detect and investigate outbreaks is critical to the response to any new threat, be it intentional or natural in origin. In the only two intentional foodborne attacks involving bacterial infectious agents in recent experience in the United States, local and state health departments detected and responded to outbreaks as a matter of course; it was not recognized at the outset that the events were intentional in origin (Torok et al., 1997; Kolavic et al., 1997). Some states have recognized that robust public health surveillance is a fundamental part of protecting the public health against both natural and intentional events, and the infusion of new resources to strengthen the response to bioterror threats has improved surveillance and response capacity in general. However, in many others, bioterror response resources have lured staff and attention away from the traditional activities of public health, leaving those systems weaker than before. Yet actual foodborne bioterror events remain remarkably rare. During the last 25 years, two have been documented, while during this same time, applying the recent Electronic Foodborne Outbreak Reporting System number of 1,200 food-borne outbreaks a year, there were an estimated 30,000 nonintentional foodborne outbreaks. Like firefighters in a firehouse restricted to arson fires, the dedicated squad will have to practice its skills in drills and table top exercises, while their less well-equipped and perhaps less well-paid colleagues put out all the fires, which must occur before the fire can be determined to be arson or not.
Enhancing Foodborne Outbreak Investigations
Despite the many constraints, the response to foodborne outbreak investigations can be improved substantially. Expanding routine subtyping to a greater number of isolates and different pathogens may help drive prevention faster and more broadly. Faster and more automated subtyping methods, including sequenced-based methods for both bacteria and viruses, need to be deployed as the cost and complexity of sequencing equipment decreases. Cluster detection can be speeded by providing the resources for swift transport and real-time testing rather than batch processing. Routine case interviews can be made more swift and standardized, and they can be integrated with control interviews to make investigation rapid. Improving the record keeping necessary for accurate tracing of foods to their sources, and increasing the skilled capacity in the state and national regulatory agencies to perform such investigations would speed that critical phase of investigations. Fingerprinting the pathogens isolated from foods and food animals in real time and linking those data with the human isolate database would make it possible to rapidly generate hypotheses about potential sources. Expanding the capacity for surveillance in other countries around the world and expanding regional and global surveillance networks to detect and investigate outbreaks can enhance the detection of foodborne threats at home and abroad (Tauxe and Hughes, 1996).
Conclusion
Enhancing foodborne surveillance, outbreak detection and response means better public health. Outbreaks will continue to occur, and people will continue to get ill, but with effective response, these unfortunate events can drive prevention. FoodNet is providing better data to measure trends and burden of illness and to determine sources of sporadic infections. PulseNet, the new network for molecular subtyping, is blurring the line between obvious outbreaks and apparently sporadic cases, and it is probing more deeply into the safety of the entire food system. Investigating and learning from the outbreaks is critical to achieving continuous improvement in food safety. As the public health system is likely to be the first responders in the event of an intentional attack, as well as for the far more frequent unintentional outbreaks, having a more robust and effective response is better for the public protection in either event.
THE ONGOING SAGA OF U.S. OUTBREAKS OF CYCLOSPORIASIS ASSOCIATED WITH IMPORTED FRESH PRODUCE: WHAT CYCLOSPORA CAYETANENSIS HAS TAUGHT US AND WHAT WE HAVE YET TO LEARN
Barbara L. Herwaldt, M.D., M.P.H.3
Centers for Disease Control and Prevention4
. . . For disease will peer up over the hedge
of health, with only its eyes showing . . .
For there will be computers
For there will be hard data and they will be hard
to understand
For the trivial will trap you and the important escape you
For the Committee will be unable to resolve the question
For there will be the arts
and some will call them
soft data
whereas in fact they are the hard data
by which our lives are lived . . .
—John Stone (Stone, 2004)
Prologue: An Ongoing Tale of Two Settings, Viewed from Multiple Perspectives
Cyclospora cayetanensis infection is endemic in many resource-poor and middle-income countries (Bern et al., 1999, 2000; Herwaldt, 2000; Lopez et al., 2003; Ortega et al., 1993). The United States, a resource-rich consumer country, has unwittingly imported this foreign, enigmatic parasite along with fresh produce that has been linked to outbreaks of cyclosporiasis. From the U.S. perspective, the unforeseen emergence of this microscopic pathogen has evolved into an ongoing tale of large foodborne outbreaks, which have entailed crossing jurisdictional borders and working with foreign governments, produce growers, and trade organizations (CDC, 1998, 2004; Herwaldt, 2000; Herwaldt et al., 1997, 1999; Ho et al., 2002; Lopez et al., 2001).
The parasite C. cayetanensis, which was christened in 1994 (Ortega et al., 1994), and the precedent-setting series of outbreaks of cyclosporiasis continue to be sources of fascination, frustration, challenges, and learning opportunities for the persons, agencies, industries, and governments they have directly or indirectly affected. The parasite and various aspects of the outbreaks (e.g., coordination of multisite investigations, interactions with foreign sources of implicated produce, regulatory responses when the mode of contamination is unknown, impacts on international trade) have been and continue to be subjects of case studies (Calvin, 2003; Jackson, 2006; Powell, 1998), including a U.S. Senate hearing in 1998 entitled The Safety of Food Imports: From the Farm to the Table: A Case Study of Tainted Imported Fruit (U.S. Senate, 1998) and this case study, which was presented in part at the IOM’s October 2005 workshop on food safety.5
This case study—in which the parasite and the U.S. outbreaks are viewed with a wide-angle lens that includes public health, societal, and historical contexts—focuses on food for thought (e.g., perspectives, principles, and issues for the public health community to ponder) rather than detailed commentary about foodborne threats to health or comprehensive review of the outbreaks of cyclosporiasis. In the text of the article, selected details about the parasite and the outbreaks are included for illustrative purposes, such as to underscore lessons learned, relearned, or yet to be learned and to highlight common themes (e.g., challenges intrinsic to emerging pathogens). Supplemental details and perspective are provided in figures (Figure 3-4, Figure 3-5) and tables (Tables 3-3 through 3-6, which may be found at the end of this article in Annexes 3-1 through 3-4). Table 3-4 and Table 3-5 represent attempts to list and dissect the elements of foodborne outbreaks and investigations to demonstrate what various ingredients add to the mix (e.g., the challenges, opportunities, and approaches if the etiologic agent is an enigmatic parasite).6

FIGURE 3-4
Generic and Cyclospora-specific challenges in bridging the chasm between disease and prevention and control. SOURCE: B. Herwaldt and D. Juranek, CDC, Division of Parasitic Diseases, April 2006 (see Acknowledgments).
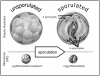
FIGURE 3-5
Sporulation of Cyclospora cayetanensis oocysts. Unsporulated and sporulated C. cayetanensis oocysts are shown in graphic illustrations (top) and photographs (bottom), as viewed by differential interference contrast (DIC) microscopy, a specialized type (more...)
TABLE 3-3
Cyclospora cayetanensis and Cyclosporiasis: Perspectives and Status as of April 2006.
TABLE 3-4
Characteristics of Foodborne Outbreaks and Investigations: Challenges, Opportunities, Approaches, and Advances, in General and Specific to Outbreaks of Cyclosporiasis Associated with Imported Fresh Produce.
TABLE 3-5
Goals of Investigations of Foodborne Outbreaks: Challenges, Opportunities, Approaches, and Advances, in General and Specific to Outbreaks of Cyclosporiasis Associated with Imported Fresh Produce.
TABLE 3-6
Factors that Have Complicated Efforts to Communicate About Foodborne Outbreaks of Cyclosporiasis.
Investigating the initial outbreaks in the 1990s would have been even more difficult than it was if Cyclospora and cyclosporiasis had emerged in the United States as complete unknowns. The fact that they did not reflects the contributions and astute observations of relatively few persons with expertise in parasitology and tropical medicine, in diverse places such as Papua New Guinea, Peru, and Nepal (Ashford, 1979; Hoge et al., 1995; Ortega et al., 1993, 1994). Their efforts culminated in fundamental scientific and medical advances, described in articles published seemingly just in the nick of time. Through studies in Peru, the confusion about the identity of the organism was resolved: it is not a species of blue-green algae (cyanobacteria); it is a coccidian parasite, the first and only species in the Cyclospora genus known to infect humans (Ortega et al., 1993, 1994). In a placebo-controlled clinical trial in Nepal, the antimicrobial combination of trimethoprim-sulfamethoxazole was demonstrated to be highly effective treatment of cyclosporiasis (Hoge et al., 1995), the first and only such therapy to have been documented (Table 3-3, Annex 3-1).
Unfortunately, the parasitologists were not prophets: the experts were as surprised as the novices by the unpredicted U.S. emergence of C. cayetanensis and by the unprecedented association between a parasite and large, common-source foodborne outbreaks. Although other enteric parasites are known to be transmissible by contaminated food, nothing remotely comparable to the widespread, recurrent outbreaks of cyclosporiasis has been documented in the United States for any other parasite. The first of the series of major eruptions of C. cayetanensis on the international scene occurred in the spring of 1996, after premonitory rumblings earlier in the decade (Herwaldt, 2000; Huang et al., 1995; Koumans et al., 1998). The eruption in 1996 took the form of a large (>1,000 reported cases7), multinational outbreak of cyclosporiasis in two countries, the United States and Canada, that was linked to a third country, Guatemala, where the epidemiologically implicated raspberries were grown (Herwaldt et al., 1997).
The modus operandi of this pathogen in the United States as the etiologic agent of outbreaks has not changed during the subsequent decade, although the repertoire of food vehicles and sources has expanded beyond raspberries from Guatemala to include assorted types of fresh produce from several middle-income countries (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3). The saga of outbreaks appears to have evolved into an interminable tome, with no end in sight. Its inscrutable chief character, C. cayetanensis, a unicellular (protozoan) parasite, continues to wreck havoc, surprise, outsmart, baffle, and bewilder us (Table 3-3, Annex 3-1). As discussed in this article, Cyclospora epitomizes the challenges intrinsic to addressing obscure pathogens that appear, seemingly out of nowhere, including how and why the challenges translate into difficulties investigating and preventing outbreaks and communicating among health professionals, the general public, and the produce industry. The scientific unknowns and political overtones are among the factors that have complicated efforts to communicate and collaborate (see Tables 3-4 through 3-6, Annexes 3-2 through 3-4).
Cyclospora, the U.S. outbreaks of cyclosporiasis, and their aftermaths have affected physical, economic, and communal health in exporter and importer nations (Calvin, 2003; Herwaldt, 2000; Jackson, 2006; Powell, 1998) (Table 3-6, Annex 3-4). The need to invest resources to investigate the outbreaks has resulted in increased recognition of and interest in this parasite and its effects on the persons, products, and places where Cyclospora infection is endemic. The extent to which the heightened awareness will stimulate long-term investments in multi-disciplinary, multilingual research activities; the research will solve remaining mysteries about this elusive pathogen, its quirky human hosts, and their micro-and macrohabitats; and the increased knowledge will be translated into wisdom, vision, and sustainable, effective, transnational prevention and control measures remains to be seen and recorded. The potential for additional scientific advances to have positive ripple effects that extend beyond Cyclospora and cyclosporiasis is high.
Challenges in Addressing Public Health Issues in General and Foodborne Outbreaks of Cyclosporiasis in Particular
Addressing public health issues, even ostensibly straightforward matters, can be difficult in part because of competing demands for scarce resources. The challenges are compounded for chronic issues such as foodborne cyclosporiasis that are associated with confounding complexities and unknowns. Figure 3-4 and the text in this section of the article represent attempts, replete with mixed metaphors, to place the challenges in perspective by depicting and describing the elements of a support structure for a public health bridge between disease and prevention and control. The goal (i.e., to bridge the chasm between disease and prevention and control), the base on which the support structure for the bridge is built (i.e., the public health infrastructure), and the societal and historical contexts for public health activities are not unique to cyclosporiasis.
Building and maintaining a structurally sound, science-grounded bridge are challenging (i.e., are uphill struggles, as depicted by the angle of the bridge). The instability and vulnerability of the base (soil) are major impediments. The infra-structure is portrayed as underground (unseen), overburdened, stretched, atrophied, eroded, and diverted. The impoverished infrastructure includes the residual resources of all types, at all tiers (e.g., local, state, federal) of the public health system. Core infrastructure constraints can be appreciated through the lens of an analogy, in which the public health system is viewed as an internal combustion engine, subject to the laws of thermodynamics (see footnote for details).8 Revitalizing the infrastructure will require commitment, concerted effort, and coordination from and among all tiers of the public health system, as well as great wisdom and vision, archetypical eroded, diverted elements.
Addressing foodborne cyclosporiasis entails adding more loads to the overburdened infrastructure in the form of the support elements for the public health bridge between disease and prevention and control. The elements are depicted as:
- a foundation stone, which symbolizes the importance of addressing fundamental constraints intrinsic to emerging pathogens; and
- pathogen and vehicle pillars (support columns), which symbolize the needs to address the superimposed challenges specific to the type of pathogen (the unique peculiarities of the coccidian parasite C. cayetanensis [Table 3-3, Annex 3-1]) and the types of food vehicles (not just fresh produce, but imported produce served in inconspicuous ways, such as garnishes [Table 3-4, Annex 3-2; Table 3-5, Annex 3-3]).
A multidimensional, complex web of interactions is depicted by the checkerboard pattern under the bridge. Although the concept of pathogen-vehicle interactions is highlighted, additional types of interactions (synergisms, antagonisms, collaborations, feedback loops) among the elements of the support structure for the bridge, the infrastructure, and society at large are germane (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3). The fact that the parasite C. cayetanensis is emerging while parasitologists are becoming an endangered species is an ironic example of a negative pathogen-infrastructure interaction.
The public health infrastructure is further stretched and strained if the challenges associated with emerging pathogens must be addressed in the context of emergencies (the extraordinary demands associated with large, multisite outbreaks [Table 3-4])—i.e., if a base of scientific knowledge about the pathogen and databases for outbreaks must be created concurrently and on the fly. If the infrastructure withstands the cumulative burden of many converging stresses, outbreak investigations, in concert with basic and applied research activities, provide opportunities to solve mysteries through scientific advances and to identify and reinforce weak elements in the public health system (Buchanan, 1997; Hall, 1997; Tauxe, 1997) (see Table 3-4, Annex 3-2; Table 3-5, Annex 3-3). The yin of outbreaks can be partially converted into yang, by translating challenges into opportunities into advancements in science and the public good.
Challenges in Addressing Emerging Pathogens, Parasites, and Cyclospora cayetanensis
Emerging Pathogens
The challenges intrinsic to emerging pathogens—particularly to newly described orphan microbes without close relations—include many constraints, all of which did or still apply, to varying degrees, to C. cayetanensis, the quintessential emerging pathogen (Table 3-3, Annex 3-1). The constraints include:
- lack of fundamental knowledge about the biology of the organism (e.g., life cycle) and therefore the epidemiology of infection (e.g., modes of transmission), which translate into difficulties predicting the behavior of the organism (e.g., responses to environmental stimuli), determining the pertinent questions to include in epidemiologic surveys, and evaluating the plausibility of competing hypotheses in outbreak investigations (Tables 3-4 through 3-6, Annexes 3-2 through 3-4);
- lack of competence and expertise among specialists (e.g., parasitologists), let alone lack of general knowledge and familiarity among other health professionals and lay persons (i.e., the little known by supposed experts is not widely known or readily accessible);
- lack of tools (e.g., analytic methods, techniques to propagate viable organisms, decontamination strategies); and
- lack of research materials (e.g., ample quantities/isolates of the organism).
The handicaps can be restated and classified in such categories as ignorance (“don’t have a clue but wish I knew”), uncertainty (“not sure and might be wrong”), unpredictability (“wouldn’t even hazard a guess”), unfamiliarity (“don’t ask me”), and unavailability (“urgently need but don’t have”), further modified by the likelihood the formidable obstacles can be overcome in the foreseeable future.
Parasites, Including Cyclospora cayetanensis
Many of these constraints apply to parasites, even to those not considered emerging pathogens per se. For example, as a broad generalization, health professionals and lay persons are less knowledgeable about and familiar with parasites than bacteria. Addressing foodborne enteric parasites—particularly protozoa, which include the coccidia C. cayetanensis and Cryptosporidium species9 (Table 3-3, Annex 3-1)—requires a paradigm shift. Although the statement that protozoa are not bacteria is a truism, the ramifications of the fact that protozoa do not behave like bacteria (e.g., do not multiply outside the host, in the environment, or in food) have been difficult to communicate.
The scientific and communication challenges posed by foodborne outbreaks of cyclosporiasis have been exacerbated by the different characteristics and behaviors of the major enteric coccidia (Table 3-3, Annex 3-1). An example of a fundamental biologic difference with epidemiologic ramifications is that Cryptosporidium oocysts (the environmental stage of coccidia) in freshly excreted stool are infective, whereas Cyclospora oocysts are not; they must mature (sporulate) outside the host to become infective, which is thought to require days to weeks under favorable environmental conditions (Table 3-3, Annex 3-1; Figure 3-5). The identified differences between Cyclospora and Cryptosporidium species in the realms of what is known about them underscore the potential risks of extrapolating from the knowns about Cryptosporidium species to fill gaps in knowledge about Cyclospora. The persistence of many enigmas about C. cayetanensis reflects the elusive search for pertinent model systems, the paucity of Cyclospora oocysts available for research purposes, and the pathogen’s short recorded history, as recounted below.
Challenges Entailed in Discovering and Classifying Cyclospora cayetanensis (1977–1994)
Reverberating Themes
Portions of the chronicle of the (re)discovery and (re)classification of the pathogen christened C. cayetanensis in 1994 are retold and dissected here to underscore recurring themes in the ongoing saga of cyclosporiasis, including common themes for emerging pathogens. Examples of such themes include the importance of:
- distinctions between signals and noise and the difficulties encountered in detecting and identifying previously undescribed, nondescript, rare signals (e.g., pathogens, outbreaks, contaminated produce) in complex mixtures of noise (e.g., stool specimens, surveillance data, salads);
- basic skills (e.g., looking, seeing, watching, waiting, counting, thinking, writing, reading);
- basic tools (e.g., microscopes for detecting oocysts and telephones for expeditious reporting of cases and potential outbreaks of cyclosporiasis).
Distinguishing Signals and Noise: Detecting Parasites Amidst the Sea of Specks in Stool Specimens
Detection and identification of protozoa, even those already known to be pathogens, by light-microscopic examination of stool specimens, is labor intensive and challenging. When viewed by light microscopy, stool specimens appear to be seas of specks; environmental samples have even more specks, lures, and pathogen imposters. Determining which of the specks are or might be parasites is difficult, especially if methods for highlighting or tagging the pertinent specks are unavailable (Table 3-3, Annex 3-1). Cyclospora oocysts (8–10 μm in diameter) can easily be missed; the oocysts in freshly excreted stool (i.e., those seen by clinical microbiologists) are unsporulated, with undifferentiated, nondescript cytoplasm and typically are shed at low levels (Table 3-3, Annex 3-1; Figure 3-5) (Herwaldt, 2000).
The Ashford and Ortega Chapters
In retrospect,10 the first three described cases of C. cayetanensis infection were detected in 1977 and 1978, by Ashford, a British parasitologist working in Papua New Guinea (Ashford, 1979). If someone without Ashford’s extraordinary eye for detail and parasitologic expertise had looked through the oculars of the microscope during “routine stool examination” of the patients’ specimens, the presence and importance of the odd, “scanty” structures in the specimens almost assuredly would have been missed or dismissed. Ashford’s discovery is all the more remarkable because techniques that facilitate detection and identification of this organism were not yet available (Table 3-3, Annex 3-1).
Explicitly listing the steps (ingredients) entailed in Ashford’s discovery is illustrative.
- He took the time to examine the stool specimens (i.e., he had to “look” to be able to “see”).
- He found/saw the “scanty” structures (unsporulated oocysts) in the sea of specks (i.e., he detected the “signal” despite the “noise”).
- He took note of them and realized that they were microbes, not debris (i.e., he both “saw” and “perceived”).
- He recognized that they were unusual (“distinct”) (i.e., he had sufficient experience to distinguish “usual/typical” from “unusual/atypical”).
- He recognized that they had features suggestive of coccidian oocysts (i.e., he had sufficient knowledge to begin to categorize the structures).
- He monitored the structures to see whether they would sporulate (i.e., he knew how to test the hypothesis that they were coccidian oocysts and had sufficient interest to do so).
- He did not yield to the temptation to discard the specimens before sporulation finally was demonstrable (at 8 days in 1 case and 11 in another).
- He recognized his limitations: he could not discern, with confidence, the number of sporozoites (i.e., two) in each of the two sporocysts in a sporulated oocyst and realized that his tally (i.e., four) might be (and was) incorrect.
- He understood the ramifications of his limitations: he could not place the organism in a genus or christen it because precise counts of the numbers of sporozoites and sporocysts, not best guesses, are required for taxonomic classification of coccidia by traditional morphologic criteria.11
- But he also recognized the importance of publishing his observations, with conservative conclusions, which he did in 1979, in the Annals of Tropical Medicine and Parasitology, in an article entitled “Occurrence of an undescribed coccidian in man in Papua New Guinea” (Ashford, 1979).
Ashford’s report (signal) about the anonymous organism he noticed was virtually unnoticed (undetected) for over a decade,12 as, presumably, the organism had been for much longer. The Ashford chapter in this chronicle raises the first in a series of laments (e.g., “if onlys” and “what ifs”). Whether and how the course of Cyclospora history would have been different, if Ashford had chosen a different title or journal for his article or had worked in an ideal world, without real-world constraints, are subjects for fairy tales rather than nonfiction; in fairy tales, pathogens would emerge with passports that included their pedigrees and profiles (e.g., personal and family names, vital statistics, travel histories), as well as high-resolution photographs of their key morphologic features.
The subsequent accomplishments of parasitologists Ortega and colleagues are also remarkable. They surmounted hurdles whose height and importance can be fully appreciated only by parasitologists. In the 1980s and early 1990s, while studying Cryptosporidium infection in Peru, they noticed what appeared to be a “big Cryptosporidium” species. Fortunately, they were not content with initial impressions (the organism is not a Cryptosporidium species), just as Ashford had not been content with best guesses (his best guess about the number of sporozoites per sporocyst would have resulted in misplacement of the microbe in the Isospora genus). Ortega and colleagues were able to demonstrate sporulation/excystation of the structures they noticed and to determine, with confidence, the morphologic features of a sporulated oocyst (Figure 3-5), which enabled them to place the organism in the Cyclospora genus and to debunk speculations about its identity (e.g., a cyanobacterium-like body).13
They described their observations in 1993, in The New England Journal of Medicine, in a seminal article entitled “Cyclospora species: A new protozoan pathogen of humans” (Ortega et al., 1993), unaware of Ashford’s report 14 years earlier in a tropical medicine journal (Ashford, 1979). Ashford read (detected) Ortega’s article in 1993 and realized that it described the organism he had correctly categorized as a coccidian parasite but had been unable to classify by genus. In 1994, in an article in which Ortega and colleagues officially christened the organism as Cyclospora cayetanensis (Table 3-3), Ashford’s contributions from back in the 1970s, from Papua New Guinea, were cited (Ortega et al., 1994).
The Incalculable Value of Cadres of Experts
The chronicle of the discovery and classification of C. cayetanensis underscores the importance of cultivating and retaining cadres of persons with specialized—sometimes seemingly old-fashioned—skills, tools, and depth and breadth of expertise: the expertise includes uncanny abilities to straddle decades, even centuries (e.g., to have historical memory and perspective but not be tethered to the past, and to be comfortable with both morphologic and molecular data); to ponder imponderables; and to see and connect microscopic dots (specks). Protecting and safeguarding brain trusts (i.e., preventing brain drains and diversions) is much easier and wiser than attempting to register and recover lost treasures. The key roles played by parasitologists, skilled in the fine arts of light microscopy and morphologic identification and classification of parasites, exemplified by their predecessor Antoni van Leeuwenhoek,14 cannot be overemphasized. Their contributions not only were prerequisites for the subsequent advances in the realm of molecular biology (Orlandi et al., 2002, 2003; Relman et al., 1996; Steele et al., 2003; Varma et al., 2003; Verweij et al., 2003) but continue to be essential (e.g., to date, molecular techniques cannot distinguish unsporulated and sporulated oocysts).
Challenges Associated with Detecting and Investigating the First Three Documented U.S. Outbreaks (1990 and 1995)
The accounts of the first three documented U.S. outbreaks of cyclosporiasis, which occurred in 1990 and 1995, underscore the understandable difficulties intrinsic to investigating outbreaks when little is understood about the etiologic agent (e.g., potentially pertinent questions and hypotheses are not considered or rigorously explored). All three of these outbreaks were detected because of unusual circumstances: stool specimens from the index case-patients were examined in clinical microbiology laboratories in which techniques that facilitate detection of Cyclospora were used routinely in parasitologic examinations (Table 3-3, Annex 3-1).
The Outbreak in Illinois in 1990: Waterborne or Foodborne?
The first documented U.S. outbreak occurred in a physicians’ dormitory in Illinois, in the summer of 1990, before the confusion about the identity of the organism had been resolved. The irresolvable uncertainty about whether the outbreak was waterborne, as it then was thought to be (Huang et al., 1995), or foodborne (Herwaldt, 2000) represents another in the series of laments related to Cyclospora. Although the outbreak was exceptional regardless of whether it was waterborne or foodborne, not knowing the mode or vehicle of transmission translates into missed opportunities to generate and explore hypotheses and to distinguish the exceptional that did occur from that which did not.
If the outbreak was waterborne (i.e., associated with fixing a water pump and refilling storage tanks in a penthouse area of the dormitory) (Huang et al., 1995), it represents the only described U.S. waterborne outbreak of cyclosporiasis, the means by which the water supply could have become contaminated are unclear, and the median incubation period (i.e., two days) was uncharacteristically short for common-source outbreaks of cyclosporiasis and enteric protozoan diseases in general (Herwaldt, 2000). If the outbreak was foodborne, it was associated with a meal in late June, the median incubation period was characteristically long (i.e., eight days [Table 3-3, Annex 3-1]), the food vehicle is unknown (details about the ingredients of food items, including a fruit salad, and food-specific attack rates were not obtained), it was the first in a long series of documented U.S. foodborne outbreaks of cyclosporiasis, and it preceded the next described foodborne outbreaks by five years.
The Outbreaks in New York and Florida in 1995: Harbingers of the Subsequent Foodborne Outbreaks?
In 1995, two potentially related outbreaks were detected: one associated with a country club in New York in May–June and the other with two social events in Florida in May (Herwaldt, 2000; Koumans et al., 1998). In retrospect, at least one if not both of these outbreaks in 1995 presaged the raspberry-associated outbreaks that were documented in 1996, 1997, 1998 (Canada), and 2000 (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3) (Herwaldt, 2000). The outbreak in New York in 1995, like the outbreak in Illinois in 1990, was initially thought to be water-borne (i.e., associated with consumption of water from coolers on a golf course). However, reevaluation of the data for the outbreak in New York, prompted by the subsequent foodborne outbreaks, indicated that the outbreak might have been associated with fruit, possibly raspberries (Herwaldt, 2000). Probably not coincidentally, New York and Florida—states with clinicians, laboratorians, and epidemiologists knowledgeable about cyclosporiasis—were the first of the total of 20 states to report cases in the multinational outbreak in 1996 linked to Guatemalan raspberries (Herwaldt, 2000).15
Evolving and Persisting Challenges Associated with the Next Era of Outbreaks of Cyclosporiasis: From Harbingers (in 1990 and 1995) to Sonic Booms (1996–2005)
The ongoing saga of cyclosporiasis has evolved from harbingers to serial, large outbreaks, detected during many of the seasons and most of the years in the subsequent decade (1996–2005). Despite the preparedness exercises with the harbinger outbreaks in 1990 and 1995, we16 knew little about the biology of Cyclospora or the epidemiology of cyclosporiasis in 1996, when confronted with a large outbreak (1,465 reported cases), which included 55 clusters of cases (i.e., 55 minioutbreaks associated with social and other events). In retrospect, the outbreak in 1996 can be viewed as the first course of what has become an ongoing curriculum of difficult challenges and learning opportunities: a progressive dinner with assorted types of fresh produce, nontraditional exports from several middle-income countries (e.g., raspberries and snow peas from Guatemala, mesclun lettuce from Peru, basil from Peru and Mexico17).
Selected details about some of the outbreaks are provided, for illustrative purposes, in the text and tables. As noted in the prologue, Table 3-4 and Table 3-5 provide matrices for listing and dissecting the ingredients of outbreaks and investigations. Examples of challenges, opportunities, approaches, societal contexts and trends, scientific advances, and lessons (re)learned, codified, and yet to be learned are included as well. The outbreaks and their impacts, particularly the series of outbreaks linked to Guatemalan raspberries, are discussed in detail in other articles and case studies (Calvin, 2003; CDC, 1998, 2004; Herwaldt, 2000; Herwaldt et al., 1997, 1999; Ho et al., 2002; Jackson, 2006; Lopez et al., 2001; Powell, 1998; U.S. Senate, 1998).
The Complementary Needs for, and Values of, “One/Few” and “Many”: Quality and Quantity
A reverberating theme in this narrative is the potential importance of “one/ few” (e.g., one oocyst detected in one stool specimen by one laboratorian; one commonality noted among a few reported cases of infection; one harbinger, clue, or outlier), which can have a positive impact (e.g., the one reported case could be the index case of an otherwise unrecognized multicluster outbreak) or a negative impact (e.g., a red herring misinterpreted as evidence). Missed oocysts and cases translate into missed outbreaks and learning opportunities and into anxious patients with un(mis)diagnosed cases of infection treated with ineffective therapies (Table 3-3, Annex 3-1; Table 3-6, Annex 3-4).
A complementary theme is the value of “many” (e.g., the unmet research need for large quantities of oocysts [Table 3-3, Annex 3-1] and the epidemiologic benefits of outbreaks with many cases and clusters of cases [Table 3-4, Annex 3-2]). Large, multicluster outbreaks, such as the outbreaks in 1996 and 1997—although challenging to investigate and coordinate and potentially devastating for the affected persons, producers, economies, and societies (Table 3-6, Annex 3-4)—can be blessings in disguise. Such outbreaks can translate into opportunities to compile compelling epidemiologic evidence to implicate food vehicles and sources (e.g., Guatemalan raspberries) and, ultimately, to learn to prevent contamination and transmission. Geographically dispersed, multicluster outbreaks provide opportunities (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3), such as:
- to circumvent limitations in knowledge about plausible modes and vehicles of transmission and to overcome limitations of individual clusters of cases (e.g., by triangulating among the clusters and events to identify the common themes in the mixtures of produce vehicles and potential sources);18
- to determine where food vehicles became contaminated (e.g., if the source is the only commonality in the distribution system of a widely dispersed vehicle);
- to generate hypotheses about modes by which the vehicle became contaminated at its source (e.g., use of contaminated agricultural water); and
- therefore, to identify and implement potentially effective control measures (i.e., to translate science into action).
Recurrent outbreaks (e.g., the series of outbreaks linked to Guatemalan raspberries), despite of and because of the challenges they pose, also provide opportunities (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3), such as:
- to (re)test hypotheses and to strengthen already compelling epidemiologic evidence (e.g., through the investigation of the raspberry-associated outbreak in the spring of 1997, which was uncannily similar to the outbreak in 1996 [Herwaldt et al., 1997, 1999]);
- to complement compelling epidemiologic evidence with confirmatory laboratory data (e.g., as was done in 2000 in the investigation of the last documented outbreak linked to Guatemalan raspberries because frozen leftovers of the implicated food item—a wedding cake—were found and analytic methods for this complex food matrix were developed [Ho et al., 2002] [Table 3-5, Annex 3-3]); and
- to assess the (in)effectiveness of control measures implemented in the past and to pause to reevaluate how to proceed (e.g., in the spring of 1998, in an inadvertent intervention trial, the United States did not import fresh Guatemalan raspberries, whereas Canada did and experienced an outbreak [CDC, 1998; Powell, 1998] [Table 3-4, Annex 3-2]).
Large and recurrent outbreaks (signals) are blessings in disguise in other ways as well: they get attention in ways that subtle signals do not. Dismissed cases (signals) in distant lands (i.e., cases not considered “our problem”) can ultimately translate into sonic booms so loud (U.S. outbreaks so large) that they cannot be ignored (Table 3-4, Annex 3-2). They serve as piercing wake-up calls about the vulnerability of our food supply in general and fresh produce in particular, including the need to bring parasites and parasitologists to the food safety and science tables. These wake-up calls can spur the growth of some good fruits, as discussed below.
Detecting and Classifying Slow-Growing Fruits
Various types of slow-growing fruits pertain to the narrative and illustrate yet again the yin and yang of the chronicle of Cyclospora. Certain principles are evident. The unexpected not only occurs but occurs in unforeseen ways and places, at unpredicted times, and with unanticipated effects. The fruits of investments in research, foreign technical assistance, the U.S. public health system, and outbreak investigations might not be evident for years, might not meet expectations (e.g., unfulfilled hopes for trickle-down benefits of investments in particular aspects of the public health infrastructure), or might not resemble the seeds that were intentionally or unwittingly planted.
We remain indebted to the astute scientists whose years of investments yielded timely fruits. Because of them we faced the outbreaks of cyclosporiasis knowing that the etiologic agent is a coccidian parasite, which slowly matures (i.e., has a long extrinsic maturation period), and knowing how to treat cases of cyclosporiasis, which can be slow to resolve (Ashford, 1979; Hoge et al., 1995; Ortega et al., 1993, 1994) (Table 3-3, Annex 3-1). The scientists did not invest in their research activities with foreknowledge that Cyclospora would emerge as it did, that the return on their investments would be large, and that U.S. investigators and patients would be among the beneficiaries. The fact that we could pluck the fruits of their years of labors should not be taken for granted.
Some of the U.S. outbreaks of cyclosporiasis, most notably the series of outbreaks associated with Guatemalan raspberries, represent a different type of slow-growing fruit of different types of labors and investments in the 1980s. Unintended fruits of good intentions to address an economic problem (i.e., to strengthen the economies of countries such as Guatemala by decreasing their reliance on traditional export crops) ripened in the 1990s, when markedly increased volumes of raspberries, a nontraditional export crop, were flown from South to North (Herwaldt, 2000; Jackson, 2006; U.S. Senate, 1998). Unintended fruits came in the form of widely dispersed outbreaks of cyclosporiasis, which had economic consequences not only for raspberry growers in Guatemala (i.e., the raspberry business essentially went out of business) but also for collaterally affected growers and suppliers of various types of fresh produce in countries including the United States (Calvin, 2003; Herwaldt, 2000; Jackson, 2006).
It would be overly simplistic to consider the good intentions in the 1980s misguided or shortsighted and to classify the fruits noted in the 1990s and thereafter as bad.19 The outbreaks and investigations have spurred the growth and maturation of various types of good fruits, with various beneficiaries. Examples of ripening good fruits (Table 3-4, Annex 3-2; Table 3-5, Annex 3-3) include the ongoing processes of:
- increasing knowledge and competence (e.g., increasing understanding of the clinical spectrum and manifestations of Cyclospora infection and developing training tools to strengthen capacity related to parasitic diseases in general and cyclosporiasis in particular); and
- developing, using, evaluating, and improving various generic and Cyclospora-specific tools (e.g., diagnostic techniques for clinical specimens, analytic methods for food and environmental samples, surveillance systems, epidemiologic survey instruments, protocols for traceback and farm investigations, control measures).
In addition, the translation of the outbreaks into piercing wake-up calls about the vulnerability of our food supply in general and fresh produce in particular has been associated with maturing fruits, with industry-wide ramifications. The outbreaks of cyclosporiasis linked to fresh produce have added to the critical mass of data from other such outbreaks (Table 3-4, Annex 3-2) to help sharpen the focus of the food-safety lens on produce vehicles in the United States and abroad. In short, good science takes time but already has yielded profits that extend beyond this particular unicellular pathogen and include science-grounded food safety guidelines and policies (e.g., “Good Agricultural Practices” for produce).
Epilogue: The Saga Is Ongoing, More Work Needs to Be Done, and the Workers and Resources Are Few
The work is not done just because the organism (i.e., C. cayetanensis) has been classified by genus and christened, one highly effective therapy for cyclosporiasis has been identified, our abilities to detect and investigate cases and outbreaks have markedly improved, and specific produce vehicles and sources have been identified in some of the outbreak investigations. Cyclospora did not gracefully exit after its dramatic entrance, nor has it fully emerged from obscurity into clarity. Despite scientific advances at the margins, Cyclospora remains largely veiled in mystery; we remain plagued by fundamental unknowns and by fundamental constraints as we seek to convert unknowns into knowns (Table 3-3, Annex 3-1). Our ability to prevent contamination of produce and thereby to prevent what we have been investigating (i.e., foodborne outbreaks) would be markedly improved by increased understanding of the biology of Cyclospora and the epidemiology of cyclosporiasis.
The lack of commitment to and the paucity of resources for the long-term investments—i.e., marathons not sprints—needed to solve the many remaining mysteries are disconcerting, especially because the ramifications of our ignorance and the collateral benefits of the advances to date are palpable. If for no other reason than self-interest (e.g., to avoid the actual and opportunity costs of investigating serial outbreaks), the need to decipher Cyclospora’s enigmatic code is evident. Cracking the code of a microbe as hardy and resilient as Cyclospora—e.g., identifying decontamination strategies that kill or remove the pathogen without damaging delicate fresh produce—almost assuredly would have far-reaching positive ripple effects for food safety.
One of the reverberating themes in the saga of this parasite is that the investments of “few” (e.g., the parasitologists’ mite) can result in large proceeds. Fortunately, some doggedly persistent scientists are continuing to poke and prod Cyclospora, despite the paucity of resources and oocysts, with the expectation that ultimately it will excyst its secrets. These resilient investigators emulate the parasite’s uncanny abilities to survive in austere microniches and to resist the pressures and stresses in harsh macroenvironments. Relatively few persons deserve the credit for the fact that Cyclospora and cyclosporiasis did not emerge in the United States as complete unknowns. In the future, which, by definition, is unknown, relatively few persons may deserve the credit for identifying and implementing an exit strategy for this inscrutable pathogen and for writing the last chapter of the chronicle of cyclosporiasis, which now appears to have no end in sight.
Acknowledgments
I am indebted to the countless persons who have contributed in various ways to the marathon investigations of the U.S. outbreaks of cyclosporiasis. I reluctantly acknowledge that Cyclospora, which has defined the seasons of more than a decade of my life, has been a worthy opponent. Special thanks to my CDC colleague, Dr. Dennis D. Juranek, for his innumerable, invaluable insights and for his artistic contributions, most notably to Figure 3-4 and Figure 3-5.
ANNEX 3-1
ANNEX 3-2
ANNEX 3-3
ANNEX 3-4
SURVEILLANCE AND INVESTIGATION OF A LARGE STATEWIDE CYCLOSPORA FOODBORNE DISEASE OUTBREAK INVOLVING AN IMPORTED STEALTH INGREDIENT
Roberta Hammond, Ph.D.20 and Dean Bodager, R.S., D.A.A.S., M.P.A.21
Florida Department of Health
Background
In mid-April 2005, a private laboratory reported a dozen cases of cyclosporiasis to the Florida Department of Health (FDH), Bureau of Epidemiology Surveillance Section. The total number of cases reported in 2004 for Florida was nine. For reporting week 14 ending April 16 (the week the positive results were received from the private lab) the average number of cyclosporiasis cases from 2003 to 2005 was 1.67. The number of cases up to week 14, 2005, was approximately 20 percent higher than normally expected (FDH, 2005a). By reporting week 17, the percent increase was 162 percent, a clear indication of a possible outbreak. Cases were reported from numerous counties with no initial apparent geographical or temporal pattern. This article will discuss the methods of the ensuing complex investigation, epidemiological findings, and recommendations for future investigations involving outbreaks of disease resulting from intentional or unintentional contaminants of the United States food supply.
Methods
Epidemiology teams were formed at the state and affected county levels of the FDH. The core statewide team consisted of epidemiologists from the Division of Disease Control and the Division of Environmental Health. A statewide lead investigator was established for this investigation using the established organizational position of Statewide Coordinator of Food and Waterborne Diseases. The statewide team also consisted of information management personnel, public communication experts, and administrators. Each assembled county health department team comprised county epidemiologists, environmental health personnel, and epidemiology nurses. Each of these teams also included regional food and waterborne disease epidemiologists and/or Florida Epidemic Intelligence Service epidemiologists. County health department administrators were also a very supportive part of the county teams. As the investigation progressed, owners and management of the numerous affected food service establishments became integral parts of the investigation, supplying the investigators with critical information pertaining to product and patrons. The FDA, the CDC Division of Parasitic Diseases, the Florida Department of Agriculture and Consumer Services, and the Department of Business and Professional Regulation also provided valuable assistance with consultation, formal traceback, investigation of food service facilities, and farm investigation activities.
Laboratory analysis of clinical specimens originated with private laboratories. Florida has had numerous previous experiences with Cyclospora outbreak investigations and, due to prior misidentification issues, understood the necessity of confirming the findings of private laboratories. Table 3-7 depicts a listing of previous outbreaks in Florida, other states, and other countries. Thus a system was set up whereby private laboratory results were sent to a single FDH coordinator. The private laboratories were also asked to send their slides to the FDH Bureau of Laboratories in Jacksonville for confirmation. Laboratory results were provided daily, sometimes up to 30 per day or more. This process also allowed for the opportunity to ensure completeness of information on cases. The FDH’s lab confirmation coordinator provided information for out-of-state cases to the CDC laboratory.
TABLE 3-7
Selected History, Cyclospora Outbreaks, and Vehicles: Florida, National, and International.
A web-based data collection system was inaugurated in order for individual county health department epidemiology staff to directly enter case information into a database for quick, real-time analysis. The web-based system is a module designed for outbreak investigations that is part of Florida’s statewide electronic reportable disease management system. The outbreak database included demographic and exposure variables and was monitored in real time by the lead investigator. Florida residents who were confirmed cases and entered into the web-based outbreak module automatically were also entered into the disease reporting system.
Communication of outbreak investigation progress and current descriptive data were electronically mailed regularly to appropriate FDH administration staff and investigation team members as well as to other state agency partners. Routine and timely updates were also posted on the FDH disease alert notification system, EpiCom, which is accessible by external partners in addition to all department staff. The CDC equivalent system, EpiX, was utilized for solicitation of out-of-state cases and communication nationwide. Press releases were also employed as needed, discussing the statewide number of cases, organism ecology, and methods of reducing risk of illness to the public.
The case definition for this outbreak investigation was a probable or confirmed case of Cyclospora infection, using the surveillance case definition, with onset since March 1, 2005, in a resident of or visitor to Florida. The FDH surveillance case defines a confirmed case as a clinically compatible case that is laboratory confirmed; the FDH defines a probable case as a clinically compatible case that is epidemiologically linked to a confirmed case.
Investigation Summary
Dates of exposure in the clustered cases ranged from March 19 to May 15, 2005. Dates of onset in the clustered cases ranged from March 24 to June 24, 2005 (see Table 3-8). Dates of onset of both sporadic and clustered cases ranged from March 1 to July 10, 2005 (see Figure 3-6). Predominant symptoms included diarrhea (78.5 percent), fatigue (64 percent), and abdominal pain (61.8 percent) (see Table 3-8). Over 75 percent of the cases were older than 40 years of age, 81 percent of the cases were Caucasian, 79 percent non-Hispanic, and 57 percent were female. Each case was asked a series of risk factor questions including a long list of various raw fruits and vegetables, other foods, and travel histories. The widespread nature of the cases and the lack of any readily apparent common food item was a strong indicator of a widely distributed food. The only weakly significant preliminary risk factors were iceberg lettuce (OR = 2.94, 95% CI 1.17–7.42; P < .02) and limes (OR = 8.54, 95% CI 1.13–64.79; P < .02). Initially all the cases appeared to be sporadic, but eventually some clusters emerged (see Table 3-8). Investigation of three of these clusters, from Pinellas, Flagler, and Sarasota Counties were used to determine the implicated food product. The Palm Beach County and the Orange County cluster investigations had inconclusive results.
TABLE 3-8
Cyclospora Clusters, Florida 2005: Range of Dates of Exposure, Dates of Onset, Confirmed and Probable Cases.
FIGURE 3-6
Epidemiology curve by week of onset, Florida 2005 Cyclospora outbreak. NOTE: The epidemiology curve is by week of onset, thus the first case of March 1 occurred during the week of February 27–March 5, 2005. SOURCE: Florida Department of Health (more...)
The first cluster to emerge was the Pinellas County cluster, associated with consuming food at chain restaurant A. In this cluster, there was a total of 42 cases (17 laboratory confirmed, 25 probable). The range of exposures was from April 1 to 2, 2005. The range of dates of onset was March 25–April 23, 2005. The implicated menu item was herb-flavored oil used for bread dipping with the following ingredients: olive oil, fresh basil, Italian parsley, rosemary, and fresh garlic (OR = 52, 95% CI 8.99–300.78; P < .0000001411 Fischers exact). During the investigation of the cluster associated with chain restaurant A, another small cluster became apparent at a different chain restaurant owned by the same company. A total of eight cases (four confirmed, four probable) was linked to this second cluster. The implicated item in the second cluster was bread dipping oil mixed with pesto. Both restaurants from different chains receive Italian parsley and fresh basil from the same distributor.
The second cluster, in Flagler County, was associated with consuming food at an independent restaurant. This cluster had a total of 20 cases (16 confirmed, 4 probable) with exposures ranging from April 1 to 12, 2005. The Flagler County cluster investigation also implicated a flavored bread dipping oil with the following ingredients: olive oil, fresh basil, fresh garlic, and parmesan cheese (OR = 27, 95% CI 2.29–534.3; P = .002).
The Sarasota County cluster is really five separate doctor’s offices whose staffs were provided catered lunches from the same independent restaurant by drug company representatives. There was an additional sporadic group associated with eating at the same independent restaurant. Exposures ranged from March 19 to April 17, 2005, and dates of onset from March 24 to April 21, 2005. While no single, statistically significant food item was identified, an ingredient can be implicated through the food histories. All five medical groups were served a lunch of meat wraps, vegetable wraps, and Greek salad, all with sun-dried tomato vinaigrette. The sporadic cases ate at the restaurant where Greek salad, Moroccan salad, and cucumber salad were on the menu. The Greek salad, meat wrap, and veggie wrap all contained sun-dried tomato vinaigrette with the following ingredients: olive oil, balsamic vinegar, sun-dried tomatoes, fresh onions, salt and pepper, and fresh basil. The inability to generate a statistically significant food is attributed to the lack of controls available for the case-control study, the suspected food ingredient being in multiple menu items and lack of recall for food histories.
A short questionnaire was administered to 35 confirmed Cyclospora cases picked at random from the sporadic outbreak cases in various areas of the state to assess fresh basil consumption habits. Five cases were selected from each of seven areas. Questions were asked pertaining to exposure to herbed green salads, basil, herbs, bruschetta, pesto, and pasta salads. There were also three questions related to visiting Italian, Thai, and gourmet restaurants that commonly serve dishes with fresh basil or fresh basil garnish. The frequencies of response to the questions included two questions that had more than 50 percent of respondents answering affirmatively. These were eating at Italian restaurants (64.7 percent) and bread dipped in oil with fresh herbs (68.8 percent). An analysis of these two variables showed significance in going to an Italian restaurant and having bread dipped in olive oil with fresh herbs. The Fisher exact value was P = .03. Eighty-one percent of the 31 cases who responded to both questions had visited an Italian restaurant where the practice of dipping bread was customary and where they ate bread in this manner. The FDH, in consultation with epidemiologists at CDC and FDA, requested a formal traceback of the fresh basil based on the significance of the findings of the three disease cluster investigations and the random case-control study.
Results
This disease outbreak was caused by Cyclospora cayetanensis, a single-celled protozoan with symptoms of watery diarrhea, nausea, loss of appetite, abdominal pain, fatigue, and weight loss. The case fatality rate is very low. The incubation period is one to seven days, usually about one week, and the ensuing illness can last anywhere from one to three weeks. Typical vehicles include raspberries, basil, lettuce, snow peas, and water. Though water has been implicated, 90 percent of outbreaks of cyclosporiasis are foodborne. Cyclosporiasis is endemic in many developing countries and is often associated with diarrhea in travelers to Asia, the Caribbean, Mexico, and Peru (Heyman, 2004).
The implicated food item in this outbreak was fresh basil imported from Peru, a widely distributed food ingredient used raw in many salads, sauces, and garnishes (Food Track Inc., 2005). It has been called a “stealth” ingredient by many because unless one knows the ingredients of a particular menu item, one might not remember having eaten it. Anecdotal evidence from a visit to the implicated farm in Peru indicates that farm conditions could have been conducive to opportunities for contamination of the basil. There was a total of 592 cases with 365 confirmed and 227 probable. The investigated illness clusters accounted for 71 confirmed and 210 probable cases (see Table 3-8). A total of 493 cases were residents of Florida with 10 cases in Canadian residents and 89 residents of other states, all having visited Florida during their exposure period. Refer to Figure 3-7 and Figure 3-8 for details on geographical distribution of cases nationwide and in Florida. All out-of-state cases were visitors to Florida who were exposed in Florida during their incubation period.
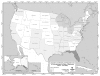
FIGURE 3-7
Laboratory-confirmed and probable cases of cyclosporiasis by state of residence, March–June 2005, Cyclospora outbreak, Florida. SOURCE: FDH (2005b).
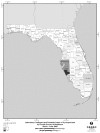
FIGURE 3-8
Laboratory-confirmed and probable cases of cyclosporiasis by Florida county of residence, March–June 2005, Cyclospora outbreak, Florida. SOURCE: FDH (2005b).
Conclusion
Due to the nature of this widely distributed stealth ingredient used raw in many common foods, this outbreak was large and diffuse and the investigation thereof was exceedingly complex, involving the entire Regional Environmental Epidemiology Strike Team, the FDH Bureau of Laboratories, staff from the Bureau of Epidemiology and all county health departments who reported cases. The FDH also collaborated with multiple partners in this outbreak investigation including private laboratories who reported cases, the Florida Department of Business and Professional Regulation, the Florida Department of Agriculture and Consumer Services, the FDA, and the CDC’s Division of Parasitic Diseases.
It can be expected that similar or more spectacular disease outbreaks will be seen in the future due to increased global distribution of foods (particularly stealth ingredients such as basil), shifts in consumption towards raw consumption of these ingredients, unusual ingredients and recipes, and the increased expectation for availability for out-of-season produce from other countries. The importation of foods from underdeveloped countries possibly with insufficient potable water supplies and processing sanitation standards is also a significant factor in these types of outbreaks. The potential for large outbreaks of this kind is great in Florida, given the large population (18 million) and the estimated annual number of visitors (74.5 million). The FDH continues to conduct surveillance for Cyclospora cases along with other emerging and reportable pathogens in order to discover outbreaks early in their occurrence so that their cause can be discovered and further spread of illness can be prevented. FDA continues its ongoing efforts in working with produce-exporting countries to ensure that produce exported to this country is safe and free from disease (FDA, 1998, 2001, 2003c, 2004; DOT and HHS, 1999).
Recommendations
Accurate laboratory analysis is critical in determining the etiological agent and scope of any foodborne outbreak. It should be noted that in this particular investigation only one clinical sample was initially misidentified as a cryptosporidium. It is imperative that public and private laboratories have the capability to accurately detect and quantify emerging pathogens and threats to our food supply as quickly as possible. These analytical and technical capabilities must include all biological, chemical, natural, and intentional threats. Public health laboratory systems need to facilitate and lead in this endeavor. Public funding allocations must reflect these high priorities for detection of food safety threats. It is also important for owners/managers and personnel at private laboratory concerns to be educated on their important role with disease surveillance and outbreak investigations and “buy in” to the investigation and critical communication processes and keep all staff apprised of this responsibility.
While the web-based system used for the data collection for cases for this outbreak investigation was somewhat helpful in rapidly collecting data from a wide geographical area, it was determined to have limited capabilities to collect control data, perform multiple variate analysis easily, and be conducive to easy manipulation of data for analysis. The limited flexibility of the design resulted in duplication of investigation and analytical efforts during some phases of the investigation. When designing and testing such elaborate systems, information technologists and software designers should include epidemiologists and other technical experts on the design and use. Scientists should also welcome and understand that programmers and data system designers should also be a part of the planning process of responding to natural and man-made biological events. Resources also need to be devoted and planned to include training of end users of these types of systems.
It should be noted that Florida was able to successfully investigate this extremely large and complex outbreak using epidemiology, nursing, laboratory, and environmental health personal within the existing organizational structures. Many resources at the county and state level in multiple scientific disciplines have been developed in the past 10–12 years that permitted this to happen. State and local municipalities who have the responsibility to conduct the surveillance, investigation, and reporting of diseases, both natural and intentional, that negatively impact public health must obtain funding to secure the human resources and develop the expertise to respond to large-scale threats to human health.
Acknowledgments
The following people provided their extensive skills and expertise with the successful investigation and reporting of this extremely large foodborne disease outbreak: Kathleen Ward, R.S., M.S.E.H., Bureau of Community Environmental Health; Mike Friedman, M.P.H., Bureau of Community Environmental Health; Robin Terzagian, Bureau of Community Environmental Health; Janet Wamnes, M.S., Bureau of Community Environmental Health; Juan Suarez, Bureau of Community Environmental Health; Carina Blackmore, Ph.D., D.V.M., Bureau of Community Environmental Health; Richard Hopkins, M.D., M.P.H., Bureau of Epidemiology; Joann Schulte, D.O., M.P.H., Bureau of Epidemiology; David Beall, Ph.D., Bureau of Laboratories; Doc Kokol, Public Information Officer; Lindsay Hodges, Public Information Officer; Maria Donnelly, M.S.P.H., Pinellas County Health Department; Sue Heller, R.N., B.S.N., Pinellas County Health Department; Hunter Zager, Pinellas County Health Department; Joe Zwissler, Pinellas County Health Department; Rick Barrett, Pinellas County Health Department; Kelly Granger, M.P.H., Hillsborough County Health Department; Aimee Pragle, M.S., Nassau County Health Department; Andre Ourso, M.P.H., Volusia County Health Department; Quintin Clark, Sarasota County Health Department; K. Eric Stutz, M.P.H., R.S., Sarasota County Health Department; Maria Teresa Bonafonte, Ph.D., Palm Beach County Health Department; Dawn Ginzl, M.P.H., Orange County Health Department; Bill Toth, M.P.H., Orange County Health Department; Barbara Herwaldt, M.D., M.P.H., CDC, Division of Parasitic Diseases.
HEPATITIS A OUTBREAKS FROM GREEN ONIONS
Beth P. Bell, M.D., M.P.H.22 and Anthony E. Fiore, M.D., M.P.H.23
Centers for Disease Control and Prevention24
During the fall of 2003, several distinct foodborne hepatitis A outbreaks occurred, including the largest such outbreak reported in the United States (Amon et al., 2005; Wheeler et al., 2005). In total, the outbreaks involved over 1,000 cases and at least three deaths. Most cases resulted from exposures in a small number of restaurants, with over 600 cases reported among patrons of a single restaurant in Pennsylvania. In each outbreak, the implicated food item was green onions imported from Mexico. As a result, the FDA imposed an import ban on green onions from the farms potentially implicated in the outbreaks (FDA, 2003a).
The outbreak investigations demonstrate a new use of molecular surveillance for hepatitis A virus (HAV) strains. Although green onions were implicated in each outbreak, the timing of the outbreaks suggested that at least two separate instances of green onion contamination occurred. This was the extent of the information that could be gleaned from the epidemiologic investigation. A more detailed understanding of the relationships among the outbreaks, gained by building on ongoing molecular surveillance, informed the traceback and affected the course of the ongoing investigations.
Features of Hepatitis A Virus Infection
Several key features of HAV infection provide the context for the outbreaks and are relevant to any consideration of prevention strategies (Fiore, 2004). After an incubation period that averages 28 days but can range from 15–50 days, HAV infection can present in a number of different ways, ranging from asymptomatic infection; to nonspecific symptoms such as nausea, abdominal pain, and fatigue; to jaundice and other classical symptoms of acute hepatitis. The likelihood of symptomatic infection and of jaundice is directly related to age; young children with HAV infection are unlikely to have a clinical illness recognizable as acute hepatitis. The period of communicability extends from about two weeks before until about one week after jaundice occurs, and HAV can be excreted in very high concentration in the stool of infected people (e.g., about 1 billion particles per gram). HAV in organic material is stable in the environment at least for a period of weeks.
Molecular Surveillance
In the investigations described here, we used molecular subtyping to characterize outbreak-related HAV strains as they became available and to explore their relationship to each other and to strains identified in the context of ongoing molecular surveillance projects in the United States and Mexico. We use nested reverse transcription polymerase chain reaction (RT-PCR) to amplify a 315 nucleotide segment at the VP1–2a junction of strains from persons with hepatitis A onset between January 2002 and August 2003, collected through the six counties comprising the Sentinel Counties Study of Acute Viral Hepatitis, and through the Border Infectious Disease Surveillance (BIDS) Project, which operates along the U.S.-Mexico border (Amon et al., 2005)
At the time the outbreak investigations began, this database included over 100 distinct sequences from over 500 individuals. Approximately 95 percent of the distinct sequences, representing 99 percent of specimens, were genotype 1A (Figure 3-9) (Amon et al., 2005). The majority of these distinct sequences formed a single cluster, in which all sequences were >96 percent similar to each other (cluster X). This cluster included sequences from all individuals identified through the BIDS project, as well as from travelers to Mexico. Particular risk factors predominated in other clusters, such as being a homosexual man or using illicit drugs (Figure 3-9).
![FIGURE 3-9. Comparison of hepatitis A viral sequences among individuals with hepatitis A from northern Mexico (Border Infectious Disease Surveillance [BIDS] Project), 2002–2003; outbreak-related surveillance, October–December 2003; and six U.S. sentinel county sites, January 2002–August 2003.](/books/NBK57087/bin/ch3f9.gif)
FIGURE 3-9
Comparison of hepatitis A viral sequences among individuals with hepatitis A from northern Mexico (Border Infectious Disease Surveillance [BIDS] Project), 2002–2003; outbreak-related surveillance, October–December 2003; and six U.S. sentinel (more...)
Outbreaks in Tennessee, North Carolina, and Georgia
The first series of outbreaks involved over 400 cases and occurred in Tennessee, North Carolina, and Georgia during August to September 2003 (Amon et al., 2005). In Tennessee and North Carolina, investigations indicated that cases were associated with one restaurant in each state, but the restaurants were unrelated to each other. Ill food service workers identified in each outbreak had illness onset concurrent with other cases, indicating that they could not have been sources of the outbreaks. Most cases in Tennessee reported an onset of illness during the first week in September. The peak in North Carolina was about a week later. In Georgia, at least three restaurant-associated clusters were identified, but information about many of the cases that shared the outbreak strain was incomplete. An epidemiologic investigation of the largest of these clusters showed that dates of onset were similar to those in North Carolina (Figure 3-10). Exposures at each restaurant occurred primarily during a 10-day period in August, and green onions were implicated as the source in case-control studies among restaurant patrons in each state. In the case-control study in Tennessee, for example, green onions were eaten by 98 percent of 57 case-patients and 46 percent of 204 control subjects (OR = 65.5, 95% CI 8.9–482.5). The epidemiologic investigations were conducted during late September and early October, and the FDA initiated traceback investigations on October 9.
FIGURE 3-10
Date of illness onset among restaurant patrons, September–November 2003; n = 590. The dotted bars represent Tennessee (TN) cases, the hatched bars North Carolina (NC) cases, the diagonal striped bars Georgia (GA) cases, and the solid bars Pennsylvania (more...)
Sera obtained from outbreak-related cases in the three states yielded two distinct outbreak strains (sequence A and B) (Figure 3-9). Sequence A was found among restaurant patrons from Tennessee, and sequence B was found among Georgia and North Carolina cases. Both sequences fell into cluster X, the same cluster of strains that also included most strains identified among persons who acquired illness in Mexico.
Pennsylvania Outbreak
In early November, as the epidemiologic investigations were winding down and the traceback investigations were underway, an outbreak in Pennsylvania occurred, eventually involving a total of more than 600 cases among patrons of a single restaurant in Beaver County, including 13 employees who became ill at the same time as patrons (Figure 3-10) (Wheeler et al., 2005). Over 80 percent of the 425 case-patients who reported eating only once at the implicated restaurant ate there between October 3 and October 6, including 67 percent who dined on October 4 or October 5. The estimated attack rate for the four-day period was 17.9 percent, including an estimated 25 percent of diners on October 4 and 29 percent of diners on October 5.
A case-control study among patrons at the restaurant included 181 cases and 89 controls. Five of the 121 menu items were associated with illness. Mild salsa was eaten by 91 percent of case-patients and 35 percent of controls (OR 19.6, 95% CI 11.0–34.9) and was the only item eaten by more than 25 percent of case-patients. Eating green onions, an ingredient in over 50 menu items, was reported by 98 percent of case-patients and 58 percent of controls and was strongly associated with illness (OR 33.3, 95% CI 12.8–86.2). The final multivariate model included age, eating mild salsa, and eating any other menu item containing green onions.
Green onions arrived at the restaurant in bundles of six to eight onions each, packed on ice in boxes. After unpacking into metal pans, they were stored in the refrigerator for up to five days. When needed, bundles were rinsed with tap water, the rubber band around the bundle was removed, and the onions were chopped using an electric dicer. After chopping, they were refrigerated in plastic containers for up to two days.
Mild salsa, the menu item most strongly associated with infection, was prepared in 40-quart batches. The restaurant prepared up to two batches each day, and stored them for up to three days in the refrigerator. Each quart contained six ounces of raw chopped green onions, equivalent to 10–16 whole green onions.
Of course a pressing question was the relationship, if any, of this outbreak to the outbreaks that had occurred earlier in the fall. Molecular surveillance results obtained to date had pointed to the likelihood of two separate instances of contamination accounting for these earlier outbreaks. The outbreak strain from cases associated with the Pennsylvania outbreak (sequence D, Figure 3-9) turned out to be distinct from but closely related to the other outbreak strains, and fell into the same cluster of Mexico-related strains. These findings established that the four geographically separate but temporally related outbreaks represented at least three distinct events.
Other States
Not all hepatitis A is foodborne, and a common question that arises in the context of many foodborne hepatitis A outbreaks is the extent to which available surveillance methods are sensitive enough such that outbreak-associated cases or small clusters can be distinguished from “background” cases. This is particularly relevant for outbreaks, such as those described here, that are associated with a distributed food item, but in which the majority of cases are associated with exposure at a restaurant. Another “first” accomplished in the context of these investigations was an improvement in the sensitivity of surveillance by incorporating molecular methods. Comparison of strains identified during the outbreak period provided evidence that some apparently “sporadic” hepatitis A cases were indeed foodborne. Specimens were requested from any cases that did not have an identified source of transmission. Of over 50 specimens submitted, a number were identical to outbreak strains (Table 3-9) (Amon et al., 2005).
TABLE 3-9
Source and Distribution of Cluster X Hepatitis A Virus Sequences, September–December 2003 (Outbreak Surveillance Specimens) and January 2002–August 2003 (Sentinel Counties and Mexico [BIDS] specimens); n = 478.
Investigation of Farms
Findings of molecular surveillance were consistent with sources in Mexico, as sequences matching each of the outbreak strains were identified from among BIDS specimens (Table 3-9). Four farms, all located in northern Mexico, potentially supplied the implicated restaurants, but no single farm could explain all four outbreaks. These traceback results were consistent with the results of sequencing—three distinct strains were identified from outbreak-associated cases in the four states.
Representatives from the FDA and CDC visited the farms in question (FDA, 2003b). The harvesting procedure included a lot of handling of the onions, which were pulled from the ground by hand, after which the outer layer was peeled off, the roots were removed, the onions were cut to a consistent size, and they were banded into bunches. Packing involved spraying bunches with chlorinated water as they passed on a conveyor belt, followed by loading into a cardboard box which was topped with chipped ice. In the distribution network, boxes generally were not handled between the farm and the restaurant destination. A number of conditions on the farm were identified as areas of concern, including poor sanitation, inadequate hand washing facilities, worker health and hygiene, the quality of the water used in the fields at packing sheds, and the ice-making process. However, no single practice or event was identified that could have explained the outbreaks.
Because HAV has no animal host, the original source of green onion contamination was a human infected with HAV and excreting the virus in stool. This fecal contamination could occur in a number of ways. Adults with contaminated hands could have touched the green onions during harvest or processing. Hepatitis A is endemic in Mexico, which means that the vast majority of the population is infected during childhood, and most adults are immune (Tanaka, 2000). Hence the majority of infections at any given time are occurring among children. Thus likely sources of contamination of hands include sewage or feces from workers’ HAV-infected children. It is also possible that HAV-infected children were present in the fields and contaminated the green onions directly. Direct contamination of the growing areas by sewage is also possible.
Discussion
The outbreaks described here were investigated rapidly and tracebacks were initiated early. However, a number of features of hepatitis A make detection and control of foodborne hepatitis A difficult, and the results of these investigations illustrate important areas of progress and remaining challenges (Fiore, 2004). Because HAV contamination of foods can be focal and the virus remains viable in the environment for months, cases can be both geographically and temporally dispersed. These investigations demonstrate the benefits of wider and faster use of molecular epidemiologic methods, both in outbreak investigations and in the context of routine hepatitis A surveillance. Viral sequencing showed that four geographically separate outbreaks that occurred in the fall of 2003 represented at least three distinct events. Sequencing activities also improved the sensitivity of surveillance to define the scope of the outbreaks, distinguish outbreak-related from nonoutbreak-related cases, and identify evidence of sporadic unrecognized foodborne transmission. Finally, viral sequencing supported the results of the first traceback investigations and accelerated control efforts related to the outbreak in Pennsylvania.
The investigations also exemplify challenges in foodborne hepatitis A outbreak response that stem from characteristics of routine hepatitis A surveillance and inherent aspects of the infection itself. Because cases reported through routine surveillance are not typically asked about foodborne exposures, the recognition of an unusual increase in the number of cases or of cases occurring among those in an unusual demographic group serves to alert authorities to begin asking about foodborne exposures as one potential common link among cases. However, even with the most rapid response and investigation of clusters of cases, the long incubation period of hepatitis A and inevitable delays in diagnosis and reporting necessitate a considerable lag time between exposure and the earliest possible detection of a foodborne outbreak. For example, the exposure that resulted in the outbreak in Pennsylvania was occurring as cases associated with the previous outbreaks were just being reported in the other states. Thus, even if a farm implicated in the earlier outbreaks had been linked to a farm implicated in the Pennsylvania outbreak, it is unlikely that even the most rapid of responses to the earlier outbreaks could have averted the subsequent outbreak in Pennsylvania.
Green onions are emerging as a potential “problem” food, having been implicated in at least two previous restaurant-associated hepatitis A outbreaks (Dentinger et al., 2001; Datta et al., 2001). The vast majority of green onions consumed in the United States are imported from Mexico, a country in which hepatitis A is endemic (Calvin et al., 2004). They require extensive handling during harvest and may be particularly difficult to clean. A pattern of focal, low-level contamination in which possibly very few bunches were contaminated, may make it difficult to detect transmission when it occurs.
The outbreak in Pennsylvania illustrates how conditions at the point of sale can amplify an outbreak. A combination of factors probably contributed to this outbreak’s size and high attack rate. The large number of diners who ate at the restaurant during the days of peak exposure were all offered mild salsa, the food item most strongly associated with illness. Preparation practices, such as rinsing green onions while they are still bundled and chopping and storage methods that allowed for cross-contamination, could also have contributed to the size of the outbreak. Because of the high concentration of HAV in stool and the likely low infectious dose, even a small amount of fecal contamination might result in many hundreds of infectious doses. Although the 2005 Food Code includes a requirement that vegetables that are not subsequently cooked be washed, it does not offer guidance about specific methods to prevent cross-contamination of produce (HHS, 2005).
Progress has been made in developing methods to detect HAV in food, including reproducible methods to detect the virus in “spiked” food samples and produce washes (Shan et al., 2005). Although theoretically attractive, there are a number of difficulties inherent in attempting to detect HAV in produce. The virus does not multiply in foods, and the concentration may be quite low. However, viral culture is not feasible, so there is a need to rely on RT-PCR techniques, which may not perform consistently in the presence of complex food mixtures. Further, RT-PCR cannot distinguish infectious HAV from noninfectious HAV RNA. Even if these technical problems were solved, HAV detection in food is unlikely to be of much practical use in the context of outbreak investigations for a number of reasons. Perhaps most important, particularly in the case of produce, is that the implicated item has almost invariably been consumed or discarded by the time illness is occurring. Further, methods are not at a level of development as of yet such that they can be scaled up to volumes needed to be reasonably sure that contamination is not present, particularly given the low infectious dose. Finally, these currently available methods take days to complete.
Perhaps most important is prevention of HAV (and other enteric pathogens) contamination of produce in the first place by preventing fecal contamination of produce on the farm. Hepatitis A is endemic in Mexico, and while the precise mechanism of transmission in the outbreaks described here could not be determined, control measures can be implemented that could prevent such outbreaks. These include ensuring that field workers are healthy and have access to adequate sanitary facilities and ensuring that water used to irrigate and rinse produce is not contaminated with feces. Children are the source of most transmission of HAV in rural communities in Mexico and much of the developing world, and children should not be present in areas where food is harvested. Reduction in HAV transmission among children in areas where produce is grown would further reduce opportunities for contamination.
REFERENCES
- Alfano-Sobsey EM, Eberhard ML, Seed JR, Weber DJ, Won KY, Nace EK, Moe CL. Human challenge pilot study with Cyclospora cayetanensis. Emerging Infectious Diseases. 2004;10(4):726–728. [PMC free article: PMC3323087] [PubMed: 15200870]
- Allos B, Moore M, Griffin P, Tauxe R. Surveillance for sporadic foodborne disease in the 21st century: The FoodNet perspective. Clinical Infectious Diseases. 2004;38(Suppl 3):S115–S120. [PubMed: 15095179]
- Amon JJ, Devasia R, Xia G, Nainan OV, Hall S, Lawson B, Wolthuis JS, Macdonald PD, Shepard CW, Williams IT, Armstrong GL, Gabel JA, Erwin P, Sheeler L, Kuhnert W, Patel P, Vaughan G, Weltman A, Craig AS, Bell BP, Fiore A. Molecular epidemiology of foodborne hepatitis A outbreaks in the United States, 2003. Journal of Infectious Diseases. 2005;192(30):1323–1330. [PubMed: 16170748]
- APHL (Association of Public Health Laboratories). A Recipe for Stronger Food Safety Testing Programs. 2003. [accessed April 12, 2006]. [Online]. Available: http://www
.aphl.org/docs /foodsafetyreport.pdf. - Ashford RW. Occurrence of an undescribed coccidian in man in Papua New Guinea. Annals of Tropical Medicine and Parasitology. 1979;73(5):497–500. [PubMed: 534451]
- Baron RJ. An introduction to medical phenomenology: I can’t hear you while I’m listening. Annals of Internal Medicine. 1985;103(4):606–611. [PubMed: 4037561]
- Bell B. Session III: The Food Supply “Threat Spectrum”—Case Studies: Cyclosporiasis in Imported Produce—Fresh Basil; Presentation at the Forum on Microbial Threats Workshop Meeting, Foodborne Threats to Health: The Policies and Practice of Surveillance, Prevention, Outbreak Investigations, and International Coordination; Washington, D.C. Institute of Medicine, Forum on Microbial Threats; Oct, 2005.
- Bern C, Hernandez B, Lopez MB, Arrowood MJ, de Mejia MA, de Merida AM, Hightower AW, Venczel L, Herwaldt BL, Klein RE. Epidemiologic studies of Cyclospora cayetanensis in Guatemala. Emerging Infectious Diseases. 1999;5(6):766–774. [PMC free article: PMC2640805] [PubMed: 10603209]
- Bern C, Hernandez B, Lopez MB, Arrowood MJ, de Merida AM, Klein RE. The contrasting epidemiology of Cyclospora and Cryptosporidium among outpatients in Guatemala. American Journal of Tropical Medicine and Hygiene. 2000;63(5–6):231–235. [PubMed: 11421369]
- Blaser MJ, Taylor DN, Feldman RA. Epidemiology of Campylobacter jejuni infections. Epidemiologic Reviews. 1983;5:157–176. [PubMed: 6357819]
- Bruzzi JF. The words count—radiology and medical linguistics. New England Journal of Medicine. 2006;354(7):665–667. [PubMed: 16481634]
- Buchanan RL. Identifying and controlling emerging foodborne pathogens: Research needs. Emerging Infectious Diseases. 1997;3(4):517–521. [PMC free article: PMC2640076] [PubMed: 9366605]
- Calvin L. Produce, food safety, and international trade: Response to U.S. foodborne illness outbreaks associated with imported produce. In: Buzby J, editor. International Trade and Food Safety: Economic Theory and Case Studies. USDA, Economic Research Service, AER-828; 2003. pp. 74–96.
- Calvin L, Avendano B, Schwentesius R. The Economics of Food Safety: The Case of Green Onions and Hepatitis A Outbreaks. 2004. [accessed February 24, 2006]. [Online]. Available: http://www
.ers.usda.gov /publications/vgs/nov04/VGS30501/ - CDC (Centers for Disease Control and Prevention). Proceedings:National Conference on Salmonellosis; March 11–13, 1964; Atlanta, Georgia: US Department of Health, Education and Welfare, Public Health Service; 1965. pp. 1–217. DHEW publication no. (PHS)1262.
- CDC. Outbreaks of Cyclospora cayetanensis infection—United States, 1996. Morbidity and Mortality Weekly Report. 1996;45(25):549–551. [PubMed: 9132573]
- CDC. Update: Outbreaks of Cyclospora cayetanensis infection—United States and Canada, 1996. Morbidity and Mortality Weekly Report. 1996;45(28):611–612. [PubMed: 8676854]
- CDC. Update: Outbreaks of cyclosporiasis—United States and Canada, 1997. Morbidity and Mortality Weekly Report. 1997;46(23):521–523. [PubMed: 9191032]
- CDC. Outbreak of cyclosporiasis—Northern Virginia-Washington, D.C.-Baltimore, Maryland, metropolitan area, 1997. Morbidity and Mortality Weekly Report. 1997;46(30):689–691. [PubMed: 9256053]
- CDC. Outbreak of cyclosporiasis—Ontario, Canada, May 1998. Morbidity and Mortality Weekly Report. 1998;47(38):806–809. [PubMed: 9776168]
- CDC. Outbreak of cyclosporiasis associated with snow peas—Pennsylvania, 2004. Morbidity and Mortality Weekly Report. 2004;53(37):876–878. [PubMed: 15385921]
- CDC. Human tuberculosis caused by Mycobacterium bovis—New York City, 2001–2004. Morbidity and Mortality Weekly Report. 2005;54(24):605–608. [PubMed: 15973241]
- CDC. Escherichia coli O157:H7 infections associated with ground beef from a U.S. military installation, Okinawa, Japan, February 2004. Morbidity and Mortality Weekly Report. 2005;54(2):40–42. [PubMed: 15660018]
- CDC. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 sites, United States, 2004. Morbidity and Mortality Weekly Report. 2005;54(14):352–356. [PubMed: 15829864]
- CDC. Foodborne Outbreak Response and Surveillance Unit [Online]. 2005. [accessed January 16, 2006]. Available: http://www
.cdc.gov/foodborneoutbreaks /outbreak_satistics /cdc_reported.htm. - CDC. FoodNet Homepage [Online]. 2006. [accessed July 7, 2006]. Available: http://www
.cdc.gov/foodnet/ - CDC. Electronic Foodborne Outbreak Reporting System (EFORS) [Online]. 2006. [accessed July 7, 2006]. Available: http://www
.cdc.gov/ - Connor BA, Reidy J, Soave R. Cyclosporiasis: Clinical and histopathologic correlates. Clinical Infectious Diseases. 1999;28(6):1216–1222. [PubMed: 10451156]
- Dalton C, Goater AD, Burt JP, Smith HV. Analysis of parasites by electrorotation. Journal of Applied Microbiology. 2004;96(1):24–32. [PubMed: 14678156]
- Datta SD, Traeger MS, Nainan OV, et al. Identification of a multi-state outbreak of hepatitis A associated with green onions using a novel molecular epidemiologic technique [abstract 896]. Program and Abstracts of the 39th Annual Meeting of the Infectious Diseases Society of America; Alexandria, VA: Infectious Disease Society of America; 2001.
- Dentinger CM, Bower WA, Nainan OV, Cotter SM, Myers G, Dubusky LM, Fowler S, Salehi ED, Bell BP. An outbreak of hepatitis A associated with green onions. Journal of Infectious Diseases. 2001;183(8):1273–1276. [PubMed: 11262211]
- de Wit M, Koopmans M, Kortbeek L, Wannet WJ, Vinje J, van Leusden F, Bartelds AI, van Duynhoven YT. SENSOR, a population-based cohort study on gastroenteritis in the Netherlands: Incidence and etiology. American Journal of Epidemiology. 2001;154(7):666–674. [PubMed: 11581101]
- Dixon BR. The prevalence and control of foodborne protozoan parasites. In: Blais BW, editor. Current Challenges in Food Microbiology. Kerala, India: Trivandrum; 2003. pp. 31–76.
- DOT, HHS (Department of the Treasury, Department of Health and Human Services). Presidential Initiative—Safety of Imported Food, Status Report, December 11. 1999. [accessed April 13, 2006]. [Online]. Available http://www
.foodsafety .gov/~dms/fs-impor.html. - Dubey JP, Thayer DW, Speer CA, Shen SK. Effect of gamma irradiation on unsporulated and sporulated Toxoplasma gondii oocysts. International Journal for Parasitology. 1998;28(3):369–375. [PubMed: 9559356]
- Eberhard ML, da Silva AJ, Lilley BG, Pieniazek NJ. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerging Infectious Diseases. 1999;5(5):651–658. [PMC free article: PMC2627716] [PubMed: 10511521]
- Eberhard ML, Ortega YR, Hanes DE, Nace EK, Do RQ, Robl MG, Won KY, Gavidia C, Sass NL, Mansfield K, Gozalo A, Griffiths J, Gilman R, Sterling CR, Arrowood MJ. Attempts to establish experimental Cyclospora cayetanensis infection in laboratory animals. Journal of Parasitology. 2000;86(3):577–582. [PubMed: 10864257]
- ERS (Economic Research Service). Economics of Foodborne Disease: Estimating the Benefits of Reducing Foodborne Disease. 2000. [accessed April 13, 2006]. [Online]. Available: http://www
.ers.usda.gov /briefing/FoodborneDisease/features .htm. - Faruque S, Sack D, Sack R, Colwell R, Takeda Y, Nair G. Emergence and evolution of Vibrio cholerae O139. Proceedings of the National Academy of Sciences. 2003;100(3):1304–1309. [PMC free article: PMC298768] [PubMed: 12538850]
- FDA (U.S. Food and Drug Administration). Multi-Year Research Strategy Under the Produce and Imported Food Safety Initiative, September. 1998. [accessed April 13, 2006]. [Online]. Available: http://www
.foodsafety .gov/~dms/fsrstrat.html. - FDA. FDA Survey of Imported Fresh Produce, January 30. 2001. [April 13, 2006]. [Online]. Available: http://www
.cfsan.fda .gov/~dms/prodsur6.html. - FDA. Import Alert #25-20: Detention without Physical Examination of Green Onions (Scallions) from Specific Firms in Mexico. 2003. [accessed April 13, 2006]. [Online]. Available: http://www
.fda.gov/ora /fiars/ora_import_ia2520.html. - FDA. FDA Statement: FDA Update on Recent Hepatitis A Outbreaks Associated with Green Onions from Mexico. 2003. [accessed February 27, 2006]. [Online]. Available: http://www
.fda.gov/bbs /topics/NEWS/2003/NEW00993.html. - FDA. FDA and CBP Bolster Safeguards on Imported Food, December 3. 2003. [accessed April 12, 2006]. [Online]. Available: http://www
.cfsan.fda .gov/~lrd/fpbtamou.html. - FDA. Prior Notice of Imported Food—Questions and Answers. 2. 2004. [accessed April 12, 2006]. [Online]. Available: http://www
.cfsan.fda .gov/~pn/pnqagui2.html. - FDH (Florida Department of Health). Bureau of Community Environmental Health, Food and Waterborne Disease Program Annual Reports. 2004. [accessed April 13, 2006]. [Online]. Available: http://www
.doh.state .fl.us/environment/community /foodsurveillance/annualreports .htm. - FDH. Bureau of Epidemiology, Weekly Morbidity Report, Week 14, Ending 4/9/05, Selected Diseases and Conditions (Confirmed Cases Only). 2005. [accessed April 13, 2006]. [Online]. Available: http://www
.doh.state .fl.us/disease_ctrl/epi /Disease_Table/2005_Weeks /dt_Week1420051.HTML. - FDH. Statewide Cyclosporiasis Outbreak, Florida, March–June 2005. 2005. Final Report January 20, 2006.
- Finelli L, Swerdlow D, Mertz K, Ragazzoni H, Spitalny K. Outbreak of cholera associated with crab brought from an area with epidemic disease. Journal of Infectious Diseases. 1992;166(6):1433–1435. [PubMed: 1431260]
- Fiore AE. Hepatitis A transmitted by food. Clinical Infectious Diseases. 2004;38(5):705–715. [PubMed: 14986256]
- Food Track Inc. Florida outbreak linked to fresh basil from a grower in Peru. Outbreak Bulletin, Cyclospora—USA (Florida)—UD#18. Food Safety & Security Alerts. 2005 Aug 20
- Fraser DW. Epidemiology as a liberal art. New England Journal of Medicine. 1987;316(6):309–314. [PubMed: 3807963]
- Frenzen P. Deaths due to unknown foodborne agents. Emerging Infectious Diseases. 2004;10(9):1536–1543. [PMC free article: PMC3320286] [PubMed: 15498153]
- Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytia-Trees E, Ribot EM, Swaminathan B. Pulsenet Taskforce. PulseNet USA—5-year update. Foodborne Pathogens and Diseases. 2006;3(1):9–19. [PubMed: 16602975]
- Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews. 1991;13:60–98. [PubMed: 1765120]
- Hall RL. Foodborne illness: Implications for the future. Emerging Infectious Diseases. 1997;3(4):555–559. [PMC free article: PMC2640088] [PubMed: 9366609]
- Havelaar A, Nauta M, Mangen MJ. Costs and Benefits of Controlling Campylobacter in the Netherlands. Bilthoven, the Netherlands: National Institute for Public Health and the Environment; 2005.
- Herikstad H, Yang S, Van Gilder T, Vugia D, Hadler J, Blake P, Deneen V, Shiferaw B, Angulo FJ. A population-based estimate of the burden of diarrhoeal illness in the United States: FoodNet, 1996–1997. Epidemiology and Infection. 2002;129(1):9–17. [PMC free article: PMC2869879] [PubMed: 12211601]
- Herwaldt BL. Cyclospora cayetanensis: A review, focusing on the outbreaks of cyclosporiasis in the 1990s. Clinical Infectious Diseases. 2000;31(4):1040–1057. [PubMed: 11049789]
- Herwaldt BL, Ackers ML. Cyclospora Working Group. An outbreak in 1996 of cyclosporiasis associated with imported raspberries. New England Journal of Medicine. 1997;336(22):1548–1556. [PubMed: 9164810]
- Herwaldt BL, Beach MJ. Cyclospora Working Group. The return of Cyclospora in 1997: Another outbreak of cyclosporiasis in North America associated with imported raspberries. Annals of Internal Medicine. 1999;130(3):210–220. [PubMed: 10049199]
- Heymann DL. Control of Communicable Diseases Manual. 18th ed. Washington, D.C: American Public Health Association; 2004. pp. 141–142.
- HHS (Department of Health and Human Services). Public Health Service Food Code [Online]. 2005. [accessed February 24, 2006]. Available: http://www
.cfsan.fda .gov/~dms/fc05-toc.html. - Ho AY, Lopez AS, Eberhart MG, Levenson R, Finkel BS, da Silva AJ, Roberts JM, Orlandi PA, Johnson CC, Herwaldt BL. Outbreak of cyclosporiasis associated with imported raspberries, Philadelphia, Pennsylvania, 2000. Emerging Infectious Diseases. 2002;8(8):783–788. [PMC free article: PMC2732521] [PubMed: 12141962]
- Hoffman RE, Greenblatt J, Matyas BT, Sharp DJ, Esteban E, Hodge K, Liang A. Capacity of state and territorial health agencies to prevent foodborne illness. Emerging Infectious Diseases. 2005;11(1):11–16. [PMC free article: PMC3294323] [PubMed: 15705316]
- Hoge CW, Shlim DR, Ghimire M, Rabold JG, Pandey P, Walch A, Rajah R, Gaudio P, Echeverria P. Placebo-controlled trial of co-trimoxazole for Cyclospora infections among travellers and foreign residents in Nepal. Lancet. 1995;345(8951):691–693. [PubMed: 7885125]
- Hoge CW, Bodhidatta L, Echeverria P, Deesuwan M, Kitporka P. Epidemiologic study of Vibrio cholerae O1 and O139 in Thailand: At the advancing edge of the eighth pandemic. American Journal of Epidemiology. 1996;143(3):263–268. [PubMed: 8561160]
- Holmes C, Chiller T. National antibiotic resistance monitoring system for enteric bacteria. Emerging Infectious Diseases. 2004;10(11):2061.
- Huang P, Weber JT, Sosin DM, Griffin PM, Long EG, Murphy JJ, Kocka F, Peters C, Kallick C. The first reported outbreak of diarrheal illness associated with Cyclospora in the United States. Annals of Internal Medicine. 1995;123(6):409–414. [PubMed: 7639439]
- IOM/NRC (Institute of Medicine/National Research Council). The science of public health surveillance, Scientific Criteria to Ensure Safe Food. Washington, D.C: The National Academies Press; 2003. pp. 28–68. [PubMed: 25057659]
- Jackson GJ. Inspection and investigation: Tools for detecting sources of food contamination and preventing illness outbreaks. In: Schweitzer G, Zali MR, Jackson G, editors. Food Safety and Foodborne Disease Surveillance Systems: Proceedings of an Iranian-American Workshop. Washington, D.C: The National Academies Press; 2006. pp. 24–54.
- Jones JL, Lopez A, Wahlquist SP, Nadle J, Wilson M. Survey of clinical laboratory practices for parasitic diseases. Clinical Infectious Diseases. 2004;38(Suppl 3):198–202. [PubMed: 15095190]
- Kassenborg H, Hedberg C, Hoekstra R, Evans MC, Chin AE, Marcus R, Vugia DJ, Smith K, Ahuja SD, Slutsker L, Griffin PM. Emerging Infections Program FoodNet Working Group. Farm visits and undercooked hamburgers as major risk factors for sporadic Escherichia coli O157:H7 infection: Data from a case-control study in 5 FoodNet sites. Clinical Infectious Diseases. 2004;38(Suppl 3):S271–S278. [PubMed: 15095199]
- Kniel KE, Lindsay DS, Sumner SS, Hackney CR, Pierson MD, Dubey JP. Examination of attachment and survival of Toxoplasma gondii oocysts on raspberries and blueberries. Journal of Parasitology. 2002;88(4):790–793. [PubMed: 12197133]
- Kolavic S, Kimura A, Simons S, Slutsker L, Barth S, Haley C. An outbreak of Shigella dysenteriae type 2 among laboratory workers due to intentional food contamination. Journal of the American Medical Association. 1997;278(5):396–398. [PubMed: 9244331]
- Koumans EH, Katz DJ, Malecki JM, Kumar S, Wahlquist SP, Arrowood MJ, Hightower AW, Herwaldt BL. An outbreak of cyclosporiasis in Florida in 1995: A harbinger of multistate outbreaks in 1996 and 1997. American Journal of Tropical Medicine and Hygiene. 1998;59(2):235–242. [PubMed: 9715939]
- Lee MB, Lee E-H. Coccidial contamination of raspberries: Mock contamination with Eimeria acervulina as a model for decontamination treatment studies. Journal of Food Protection. 2001;64(11):1854–1857. [PubMed: 11726175]
- LeFebvre B, Diarra M, Giguere K, Roy G, Michaud S, Malouin F. Antibiotic resistance and hypermutability of Escherichia coli O157 from feedlot cattle treated with growth-promoting agents. Journal of Food Protection. 2005;68(11):2411–2419. [PubMed: 16300081]
- Lopez AS, Dodson DR, Arrowood MJ, Orlandi PA, da Silva AJ, Bier JW, Hanauer SD, Kuster RL, Oltman S, Baldwin MS, Won KY, Nace EM, Eberhard ML, Herwaldt BL. Outbreak of cyclosporiasis associated with basil in Missouri in 1999. Clinical Infectious Diseases. 2001;32(7):1010–1017. [PubMed: 11264028]
- Lopez AS, Bendik JM, Alliance JY, Roberts JM, da Silva AJ, Moura IN, Arrowood MJ, Eberhard ML, Herwaldt BL. Epidemiology of Cyclospora cayetanensis and other intestinal parasites in a community in Haiti. Journal of Clinical Microbiology. 2003;41(5):2047–2054. [PMC free article: PMC154728] [PubMed: 12734247]
- McQuiston J, Parrenas R, Ortiz-Rivera M, Gheesling L, Brenner F, Fields P. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. Journal of Clinical Microbiology. 2004;42(5):1923–1932. [PMC free article: PMC404605] [PubMed: 15131150]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerging Infectious Diseases. 1999;5(5):607–625. [PMC free article: PMC2627714] [PubMed: 10511517]
- Novotny TE. US Department of Health and Human Services: A need for global health leadership in preparedness and health diplomacy [editorial] American Journal of Public Health. 2006;96(1):11–13. [PMC free article: PMC1470430] [PubMed: 16317195]
- Nuorti J, Niskanen T, Hallanvuo S, Mikkola J, Kela E, Hatakka M, Fredriksson-Ahomaa M, Lyytikainen O, Siitonen A, Korkeala H, Ruutu P. A widespread outbreak of Yersinia pseudotuberculosis O:3 infections from iceberg lettuce. Journal of Infectious Diseases. 2004;189(5):766–774. [PubMed: 14976592]
- Orlandi PA, Chu D-MT, Bier JW, Jackson GJ. Parasites and the food supply. Food Technology. 2002;56(4):72–81.
- Orlandi PA, Carter L, Brinker AM, da Silva AJ, Chu D-M, Lampel KA, Monday SR. Targeting single-nucleotide polymorphisms in the 18S rRNA gene to differentiate Cyclospora species from Eimeria species by multiplex PCR. Applied and Environmental Microbiology. 2003;69(8):4806–4813. [PMC free article: PMC169076] [PubMed: 12902274]
- Ortega YR, Sterling CR, Gilman RH, Cama VA, Diaz F. Cyclospora species: A new protozoan pathogen of humans. New England Journal of Medicine. 1993;328(18):1308–1312. [PubMed: 8469253]
- Ortega YR, Gilman RH, Sterling CR. A new coccidian parasite (Apicomplexa: Eimeriidae) from humans. Journal of Parasitology. 1994;80(4):625–629. [PubMed: 8064531]
- Ortega YR, Nagle R, Gilman RH, Watanabe J, Miyagui J, Quispe H, Kanagusuku P, Roxas C, Sterling CR. Pathologic and clinical findings in patients with cyclosporiasis and a description of intracellular parasite life-cycle stages. Journal of Infectious Diseases. 1997;176(6):1584–1589. [PubMed: 9395371]
- Powell D. Risk-based regulatory responses in global food trade: Guatemalan raspberry imports into the U.S. and Canada, 1996–1998. Conference on Science, Government, and Global Markets: The State of Canada’s Science-Based Regulatory Institutions, Carleton Research Unit of Innovation, Science and Environment; Ottawa. October 1–2, 1998; 1998. pp. 1–15.
- Rangel J, Sparling P, Crowe C, Griffin P, Swerdlow D. Epidemiology of Escherichia coli O157: H7 outbreaks, United States, 1982–2002. Emerging Infectious Diseases. 2005;11(4):603–609. [PMC free article: PMC3320345] [PubMed: 15829201]
- Redelmeier DA. The cognitive psychology of missed diagnoses. Annals of Internal Medicine. 2005;142(2):115–120. [PubMed: 15657159]
- Relman DA, Schmidt TM, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. Journal of Infectious Diseases. 1996;173(2):440–445. [PubMed: 8568307]
- Robbins JA, Sjulin TM, Rasmussen HP. Scanning electron microscope analysis of drupelet morphology of red raspberry and related Rubus genotypes. Journal of the American Society of Horticultural Science. 1988;113(3):474–480.
- Sapers GM. Efficacy of washing and sanitizing methods for disinfection of fresh fruit and vegetable products. Food Technology and Biotechnology. 2001;39(4):305–311.
- Sathyanarayanan L, Ortega Y. Effects of pesticides on sporulation of Cyclospora cayetanensis and viability of Cryptosporidium parvum. Journal of Food Protection. 2004;67(5):1044–1049. [PubMed: 15151249]
- Schlech WF III, Lavigne PM, Bortolussi RA, Allen AC, Haldane EV, Wort AJ, Hightower AW, Johnson SE, King SH, Nicholls ES, Broome CV. Epidemic listeriosis—evidence for transmission by food. New England Journal of Medicine. 1983;308(4):203–206. [PubMed: 6401354]
- Shan XC, Wolffs P, Griffiths MW. Rapid and quantitative detection of hepatitis A virus from green onion and strawberry rinses by use of a real-time reverse transcriptase-PCR. Applied Environmental Microbiology. 2005;71(9):5624–5626. [PMC free article: PMC1214659] [PubMed: 16151164]
- Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. Journal of Food Protection. 2004;67(10):2342–2353. [PubMed: 15508656]
- Slutsker L, Evans MC, Schuchat A. Listeriosis. In: Scheld W, Craig W, Hughes J, editors. Emerging Infections. Vol. 4. Washington, D.C: American Society for Microbiology Press; 2000. pp. 83–106.
- Sobel J, Khan AS, Swerdlow DL. Threat of a biological terrorist attack on the US food supply: The CDC perspective. Lancet. 2002;359(9309):874–880. [PubMed: 11897303]
- Steele M, Unger S, Odumeru J. Sensitivity of PCR detection of Cyclospora cayetanensis in raspberries, basil, and mesclun lettuce. Journal of Microbiology Methods. 2003;54(2):277–280. [PubMed: 12782383]
- Sterling CR, Ortega YR. Cyclospora: An enigma worth unraveling. Emerging Infectious Diseases. 1999;5(1):48–53. [PMC free article: PMC2627696] [PubMed: 10081671]
- Stone J. “Gaudeamus Igitur” [poem], Stone, Music from Apartment 8: New and Selected Poems. Baton Rouge: Louisiana State University Press; 2004.
- Swaminathan B, Gerner-Smidt P, Ng LK, Lukinmaa S, Kam KM, Rolando S, Gutierrez EP, Binsztein N. Building PulseNet International: An interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathogens and Diseases. 2006;3(1):36–50. [PubMed: 16602978]
- Tanaka J. Hepatitis A shifting epidemiology in Latin America. Vaccine. 2000;18(Suppl 1):S57–S60. [PubMed: 10683550]
- Tappero J, Schuchat A, Deaver K, Mascola L, Wenger J. Reduction in the incidence of human listeriosis in the United States: Effectiveness of prevention efforts? Journal of the American Medical Association. 1995;273(14):1118–1122. [PubMed: 7707600]
- Tauxe RV. Emerging foodborne diseases: An evolving public health challenge. Emerging Infectious Diseases. 1997;3(4):425–434. [PMC free article: PMC2640074] [PubMed: 9366593]
- Tauxe RV. Emerging foodborne pathogens. International Journal of Food Microbiology. 2002;78(1–2):31–41. [PubMed: 12222636]
- Tauxe RV. Session II: The Food Supply “Threat Spectrum”; Presentation at the Forum on Microbial Threats Workshop Meeting, Foodborne Threats to Health: The Policies and Practices of Surveillance, Prevention, Outbreak Investigations, and International Coordination; Washington, DC. Institute of Medicine, Forum on Microbial Threats; Oct 25, 2005.
- Tauxe RV, Hughes JM. International investigation of outbreaks of foodborne disease. British Medical Journal (Clinical Research Ed.). 1996;313(7065):1093–1094. [PMC free article: PMC2352411] [PubMed: 8916684]
- Torok TJ, Tauxe RV, Wise RP, Livengood JR, Sokolow R, Mauvais S, Birkness KA, Skeels MR, Horan JM, Foster LR. A large community outbreak of salmonellosis caused by intentional contamination of restaurant salad bars. Journal of the American Medical Association. 1997;278(5):389–395. [PubMed: 9244330]
- Treadway K. Heart sounds. New England Journal of Medicine. 2006;354(11):1112–1113. [PubMed: 16540610]
- U.S. Senate, Permanent Subcommittee on Investigations of the Committee on Governmental Affairs. The Safety of Food Imports: From the Farm to the Table: A Case Study of Tainted Imported Fruit. 105th Cong., 2nd Sess., Senate Hearing 105–516 (Part II); July 9, 1998. 1998.
- Varma M, Hester JD, Schaefer FW, Ware MW, Lindquist HDA. Detection of Cyclospora cayetanensis using a quantitative real-time PCR assay. Journal of Microbiology Methods. 2003;53(1):27–36. [PubMed: 12609720]
- Verweij JJ, Laeijendecker D, Brienen EAT, van Lieshout L, Polderman AM. Detection of Cyclospora cayetanensis in travellers returning from the tropics and subtropics using microscopy and real-time PCR. International Journal of Medical Microbiology. 2003;293(2–3):199–202. [PubMed: 12868656]
- Voetsch A, Angulo F, Rabatsky-Ehr T, Shallow S, Cassidy M, Thomas SM, Swanson E, Zansky SM, Hawkins MA, Jones TF, Shillam PJ, Van Gilder TJ, Wells JG, Griffin PM. Emerging Infections Program FoodNet Working Group. Laboratory practices for stool-specimen culture for bacterial pathogens, including Escherichia coli O157:H7 in the FoodNet sites, 1995–2000. Clinical Infectious Diseases. 2004;38(Suppl 3):S190–S197. [PubMed: 15095189]
- Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV. Emerging Infections Program FoodNet Working Group. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clinical Infectious Diseases. 2004;38(Suppl 3):S127–134. [PubMed: 15095181]
- Vogt DU. Food Safety Issues in the 109th Congress. Washington, D.C: Library of Congress, Congressional Research Service; 2005.
- Wan J, Wilcock A, Coventry MJ. The effect of essential oils of basil on the growth of Aeromonas hydrophila and Pseudomonas fluorescens. Journal of Applied Microbiology. 1998;84(2):152–158. [PubMed: 9633630]
- Wheeler JG, Sethi D, Cowden JM, Wall PG, Rodrigues LC, Tompkins DS, Hudson MJ, Roderick PJ. Study of infectious intestinal disease in England: Rates in the community, presenting to general practice, and reported to national surveillance. The Infectious Intestinal Disease Study Executive. British Medical Journal. 1999;318(7190):1046–1050. [PMC free article: PMC27838] [PubMed: 10205103]
- Wheeler C, Vogt TM, Armstrong GL, Vaughan G, Weltman A, Nainan OV, Dato V, Xia G, Waller K, Amon J, Lee TM, Highbaugh-Battle A, Hembree C, Evenson S, Ruta MA, Williams IT, Fiore AE, Bell BP. An outbreak of hepatitis A associated with green onions. New England Journal of Medicine. 2005;353(9):890–897. [PubMed: 16135833]
- Wick L, Qi W, Lacher D, Whittam T. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. Journal of Bacteriology. 2005;187(5):1783–1791. [PMC free article: PMC1064018] [PubMed: 15716450]
- Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, Okamoto H. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. Journal of General Virology. 2003;84(Pt 9):2351–2357. [PubMed: 12917455]
- Zhang X, McDaniel A, Wolf L, Keusch G, Waldor M, Acheson D. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. Journal of Infectious Diseases. 2000;181(2):664–670. [PubMed: 10669353]
Footnotes
- 1
Captain, U.S. Public Health Service; Chief of Foodborne and Diarrheal Diseases Branch, Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases.
- 2
The findings and conclusions in this manuscript have not been formally disseminated by CDC and should not be construed to represent any agency determination or policy.
- 3
Medical Epidemiologist in the Division of Parasitic Diseases.
- 4
The findings and conclusions in this manuscript have not been formally disseminated by the CDC and should not be construed to represent any agency determination or policy.
- 5
Forum on Microbial Threats. 2005 (October 25–26). “Foodborne Threats to Health: The Practice and Policies of Surveillance, Prevention, Outbreak Investigations, and International Coordination,” Washington, D.C.
- 6
Table 3-4 and Table 3-5, which are complementary, have the same column headings—i.e., outbreaks/investigations in general, the etiologic agent C. cayetanensis, fresh produce vehicles, and foreign sources. In Table 3-4, which focuses on the characteristics of outbreaks/investigations, the row headings distinguish outbreaks/investigations in general and outbreaks with various characteristics (i.e., large, multisite, concurrent, recurrent, seasonal, and associated with high attack rates). In Table 3-5, which focuses on the goals of outbreak investigations, the row headings distinguish the processes of identifying food vehicles, their sources, sites/modes of contamination, and control measures. Additional perspectives about the scientific and communication challenges highlighted in Table 3-4 and Table 3-5 are provided in Table 3-3 and Table 3-6, respectively.
- 7
Although the true magnitude of the outbreak in North America in 1996 is unknown, 1,465 cases of cyclosporiasis were reported (including 1,270 [87 percent] in the United States, in 20 states and the District of Columbia), which dwarfed the total number of cases previously documented worldwide (Herwaldt et al., 1997). In the raspberry-associated outbreak in 1997, the case count was 1,012 (including 981 [97 percent] in the United States, in 17 states and the District of Columbia) (Herwaldt et al., 1999). The total number of reported cases in the series of outbreaks in North America from 1995–2005 exceeds 4,000. The admonition to remember that cases occur in real persons bears repeating (Baron, 1985). As expressed by a physician to the medical students she mentors: “. . . you have not finished your work until you have taken care of the patient, not just the problem” (Treadway, 2006).
- 8
Within the public health system (engine), resources (internal energy) are constantly being consumed (e.g., scarce financial and personnel resources are becoming scarcer), regardless of whether the system is idling or moving an agenda and whether the direction of the movement is forward or backward. Although resources can be converted from one type or state to another (e.g., from dollars to devices and from federal to state programs), as well as redistributed, recycled, and diverted, they cannot be created de novo; acquisition of new resources requires infusions from, or collaborations with, other systems. The positive spin on the need for outside input is that it provides opportunities for invaluable perspective and reality checks.
- 9
In this article, the terminology “Cryptosporidium species” refers to C. hominis/parvum.
- 10
Some aspects of the Cyclospora chronicle are clear only if viewed through the retrospectoscope, which underscores the importance of reevaluating conclusions (e.g., from ongoing and prior outbreak investigations [Table 3-4]), as additional data and insights become available.
- 11
The miscount, although understandable (i.e., because of the configuration of the crescent-shaped sporozoites [Figure 3-5]), was not a minor matter. The difference between two sporozoites in each of two sporocysts and four sporozoites per sporocyst represents the difference between the Cyclospora and Isospora genera, respectively. Cryptosporidium species have four naked sporozoites (no sporocysts).
- 12
Searching the medical literature for potentially relevant articles about anonymous organisms is difficult.
- 13
Yet another twist in the tale was reported in the mid 1990s. Phylogenetic analyses, based on one gene (i.e., the small subunit ribosomal RNA gene), showed that C. cayetanensis is as closely related to some species of a different genus of coccidia—namely, the Eimeria genus—as some Eimeria species are to each other (Herwaldt, 2000; Relman et al., 1996). The biologic implications of this finding are unknown, but the ramifications for testing environmental samples are clear. Although, to date, no Eimeria species have been shown to be human pathogens, because they are prevalent in animals, environmental samples should be analyzed with techniques that distinguish C. cayetanensis from Eimeria species, not just other Cyclospora species. By traditional taxonomic (morphologic) criteria, Eimeria oocysts have four sporocysts (Cyclospora oocysts have two), each of which contains two sporozoites.
- 14
Antoni van Leeuwenhoek (1632–1723), Dutch microscopist extraordinaire.
- 15
The positive ripple effects of training received by one person who trains others can continue for years. In Florida, microbiology staff in a community hospital had dutifully examined stool specimens for Cyclospora ever since a training course in the spring of 1992 (i.e., before the organism had been christened), despite detecting only two cases of infection, in persons returning from overseas travel, during the ensuing three years (Koumans et al., 1998). In June 1995, the laboratory staff detected and reported the index cases of the outbreak in 1995. The staff subsequently trained laboratorians from other hospitals in the county and a state laboratory to examine stool specimens for Cyclospora. The state of Florida added cyclosporiasis to its list of reportable diseases in July 1995. Laboratory and epidemiologic staff in Florida continue to play central roles in detecting and investigating outbreaks of cyclosporiasis associated with imported fresh produce (e.g., mesclun lettuce from Peru in 1997 and basil from Peru in 2005) (Herwaldt, 2000; also see the section in this chapter by R. Hammond and D. Bodager).
- 16
The words we and our are used to include all potentially relevant persons and agencies, without any explicit or implicit focus on the author’s roles.
- 17
The examples are those that can be provided with confidence (i.e., these vehicles and sources were definitively implicated in investigations of U.S. outbreaks). The evidence for other vehicles and sources (e.g., blackberries from Guatemala, in various years including 1999, and raspberries from Chile, in an outbreak during late December 2001 through early January 2002) was not definitive because of the typical types of constraints (Herwaldt, 2000; Jackson, 2006). For example, only one cluster of cases was documented for the outbreak in the winter of 2001/2002 (i.e., no opportunities to triangulate among clusters were available); the evidence that the raspberries that were eaten—which were grown in Chile—were the vehicle of infection was suggestive but not definitive.
- 18
Epidemiologic and traceback investigations of produce-associated outbreaks would be greatly simplified, if meals and food items included a maximum of one type of fresh produce, if aliquots of such items were frozen for potential future analytic testing, and all pertinent receipts were saved.
- 19
In 2004, the saga of cyclosporiasis circled back to Guatemala, when an outbreak was linked to snow peas, another nontraditional export crop (CDC, 2004).
- 20
Statewide Coordinator, Food and Waterborne Disease Program, Bureau of Community Environmental Health.
- 21
Central Regional Environmental Epidemiologist, Food and Waterborne Disease Program, Bureau of Community Environmental Health.
- 22
Division of Viral Hepatitis.
- 23
Division of Viral Hepatitis.
- 24
The findings and conclusions in this manuscript have not been formally disseminated by the CDC and should not be construed to represent any agency determination or policy.
- OVERVIEW
- THE BURDEN OF ILLNESS ASSOCIATED WITH FOODBORNE THREATS TO HEALTH, AND THE CHALLENGE OF PREVENTION
- THE ONGOING SAGA OF U.S. OUTBREAKS OF CYCLOSPORIASIS ASSOCIATED WITH IMPORTED FRESH PRODUCE: WHAT CYCLOSPORA CAYETANENSIS HAS TAUGHT US AND WHAT WE HAVE YET TO LEARN
- ANNEX 3-1
- ANNEX 3-2
- ANNEX 3-3
- ANNEX 3-4
- SURVEILLANCE AND INVESTIGATION OF A LARGE STATEWIDE CYCLOSPORA FOODBORNE DISEASE OUTBREAK INVOLVING AN IMPORTED STEALTH INGREDIENT
- HEPATITIS A OUTBREAKS FROM GREEN ONIONS
- REFERENCES
- Investigating Foodborne Threats - Addressing Foodborne Threats to HealthInvestigating Foodborne Threats - Addressing Foodborne Threats to Health
Your browsing activity is empty.
Activity recording is turned off.
See more...