Featured Resource: BLAST+, All New BLAST Available on Web Service and for Download
The all-new NCBI BLAST+, built with the NCBI C++ toolkit, is now live on the NCBI Web service and is available for download from the BLAST area of the ftp site. BLAST+ features improved integration between the Web service and standalone package including the ability to save and use search strategies developed on the Web in a local installation. Also featured is a logical correspondence between the local binaries and the functions apparent on the Web (blastn, blastp, tblastn, tblastx, psiblast, and rpsblast). The new binaries offer improved performance, and new, more flexible formatting options. Another improvement is that the separate network BLAST client and BLAST 2 Sequences software are no longer needed as they are incorporated as options in the program binaries.
Web Service
Important changes to the Web service include the incorporation of the BLAST 2 Sequences service on the main BLAST submission forms (described in the December 2008 issue of NCBI News), changes to the output format, and new options for downloading results and search strategies.
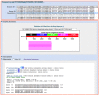
Figure 1 shows the new output for a multi-sequence protein BLAST search. Results for searches with multiple sequences are easier to read in the new format as the results are displayed one-at-a-time with the results from each query accessible from the “Results for …” pull-down list. In addition, sequences with no hits can be quickly identified in the pull-down list by grayed-out titles.
The new output format features expandable / collapsible sections of the BLAST results (Graphic Summary, Conserved Domains, Descriptions, and Alignments) that allow selecting only the most relevant sections of a particular output for display. There are also expandable Formatting and Download options sections on the output page. These new options are more convenient than the previous separate Formatting page, and the download options now provide a direct way to save BLAST results as text, structured formats (XML or ASN.1), or as the hit table tabular format. In addition, the hit table is now available in a comma separated value (CSV) format that can be directly opened with standard spreadsheet applications.
Standalone BLAST Package
The BLAST+ package is available for a variety of computer platforms from the NCBI ftp site:
ftp.ncbi.nih.gov/blast/executables/blast+/LATEST
There are a number of important differences between BLAST+ and the traditional standalone BLAST package. The most apparent is the separation of the blastall program into individual binaries by BLAST search function and the replacement of other traditional programs. Table 1 presents a partial correspondence between the old and the new package programs.
Table 1
A partial list of corresponding programs in traditional BLAST and the new BLAST+.
The new programs also feature long-form command line options instead of the traditional single letter options, making the options easier to remember and providing more flexibility for additional options in later releases. The box below shows how the same search – blastn against the NCBI nucleotide database (nt) – would be written in traditional BLAST and BLAST+. The BLAST+ command line closely parallels the way the same search would be conducted on the Web service: select the nucleotide form from the Basic BLAST section of the BLAST Homepage – the equivalent of using the blastn binary, choose blastn from the Program Selection portion of the submission form – the equivalent of using the task blastn.
Traditional BLAST:
blastall –i input.seq –d nt –p blastn –e 0.001 –o output
BLAST+
blastn –query input.seq -db nt –task blastn –evalue 0.001 –out output
New Options
New standalone options for BLAST+ include tasks, search strategies, and custom output formats.
The task option manages the different aspects of nucleotide searches for the blastn binary (blastn, megablast, discontiguous megablast) but also offers pre-set conditions for short sequence searches (-task blastn-short, -task blasp-short).
Another of the completely new features in the BLAST+ package is the ability to import and export search strategies as mentioned above for Web BLAST. In the standalone package this is managed through the –export_search_strategy and –import_search_strategy command line switches. This allows reproducible exchange of search parameters between the NCBI Web service and the local installation. For greater flexibility, a different query and database can also be specified when using a saved search strategy.
Custom outputs are available now for the BLAST tabular and comma-separated formats. These offer a large range of combinations of statistics, locations, and identifiers. Custom output formats are managed through format specifiers passed to the -outfmt option in the search program. More detailed information on these and other options for any of the standalone program is available by using the –help option on the command line of the program.
Replacement of BLAST 2 Sequences and the Netblast client
The BLAST+ package now includes the ability to compare two or more sequences to each other in each of the search programs. Using the –subject option instead of the –db will cause any of the search programs to behave as a BLAST 2 sequences program. This eliminates the need for the BLAST 2 sequences utility (bl2seq) included in the traditional BLAST package. As with the other changes this now makes the standalone package more similar to the Web version where the BLAST 2 sequences option is available for all Basic BLAST searches.
The BLAST+ package can also function as a network client to the NCBI Web BLAST service by specifying the –remote option on the command line of one of the search programs. This offers a more powerful option for submitting small to medium sized batches of sequences than the older netblast client (blastcl3) available as a separate distribution on the NCBI ftp site.
Other Improvements and Additional Help
Other important improvements in BLAST+ include performance enhancements using query splitting, the ability to use pre-indexed databases for megablast as well as new abilities and options for the BLAST database applications makeblastdb and blastdbcmd. For additional information on any of these improvements and help on using and installing BLAST+ see the BLAST Command Line Applications User Manual available on the NCBI Bookshelf.
www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpblast&part=CmdLineAppsManual
Summary
The new BLAST+ suite of programs offers more flexible options, enhanced performance, and improved integration with the NCBI Web BLAST services compared with the C toolkit BLAST. BLAST+ will continue to evolve and improve as the primary sequence similarity search tool at the NCBI and the most widely used similarity search tool in the world.
New Databases and Tools
Bookshelf
The Bookshelf has added two new books: Animal Models of
Cognitive Impairment and Baculovirus Molecular Biology. Books can be found at: www.ncbi.nlm.nih.gov/sites/entrez?db=Books.
Microbial Genomes
Sixteen finished microbial genomes were released between November 17 and December 17. The original sequence data files submitted to GenBank/EMBL/DDBJ can be found at: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are available via FTP at: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 169.0 is available via web and FTP. Release 169.0 includes information as of December 11, 2008.
A new linetype, DBLINK, will be implemented in GenBank files beginning with the February 2009 release. More information can be found in Section 1.4.1 of the GenBank Release Notes. Also, GenBank ‘index’ files are now provided without EST content, and without most GSS content. See Section 1.3.12 of the release notes for further details. Release notes can be found on the ftp site at: ftp.ncbi.nih.gov/genbank/gbrel.txt
NCBI is considering ceasing support for the index files. Affected users are encouraged to review Section 1.3.2 of the release notes and provide feedback to the GenBank newsgroup or the NCBI service desk at: vog.hin.mln.ibcn@ofni .
Updates and Enhancements
PubMed DocSum
The “Gene Sensor” is a new feature seen on PubMed results pages that provides definitive information for a gene. If a PubMed search is performed with a Gene Symbol, a Gene Information box appears on the results page with a brief description of the gene along with a link to the Entrez Gene database.
The PubMed Document Summary (DocSum) page has also been updated to provide more emphasis on an article’s title rather than its authors. These new features are part of an effort to improve the quality of search results and promote the discovery of new information.
Sequin Version 9.0
Sequin version 9.00 for Macintosh, PC/Windows, and Unix computers is available from the NCBI ftp site: ftp.ncbi.nih.gov/sequin. Feature and qualifier dialogs have been updated to comply with the latest version of the International Sequence Database Collaboration (INSDC) Feature Documentation.
PubMed Entrez Utilities
Updated 2009 NCBI PubMed Document Type Definitions (DTDs) are available from the Entrez DTD page at: eutils.ncbi.nlm.nih.gov/entrez/query/DTD/index.shtml. DTD changes for the 2009 production year are noted in the Revision Notes section near the top of each DTD.
HomoloGene
HomoloGene release 62 is available via web and ftp. This new release includes an improved approach for predicting putative paralogs. The HomoloGene website is: www.ncbi.nlm.nih.gov/homologene/. The website has a “Tip of the Day” feature that provides information on designing more successful searches.
Entrez Gene
A few changes will be seen in the Entrez Gene database in the coming weeks. An option to sort by chromosome will present gene records first alphabetically by organism name, then numerically by chromosome, and finally numerically by the start position on the chromosome. This chromosome information will now appear in the Document Summaries returned by searches in Gene.
Different gene-to-gene relationships will soon be reported including a “Related functional gene” heading and link within the Entrez Gene full report. The functional gene will have a link to related pseudogenes in the “General gene information” section.
A new preferred symbol field contains the preferred symbol for a gene. The alias for this field is [PREF].
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
Publication Details
Author Information and Affiliations
Publication History
Created: December 15, 2008.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Cooper P, Lipshultz D. NCBI News, January 2009. 2008 Dec 15. In: NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.