NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
The biological sequence information that builds the foundation of NCBI's databases and curated resources comes from many sources. How are these data managed and processed once they reach NCBI? This chapter discusses the flow of sequence data, from the management of data submission to the generation of publicly available data products.
Overview
The central dogma of molecular biology asserts that sequences flow from DNA to RNA to protein. In Entrez, DNA and RNA sequences are retrieved together as nucleotides and then integrated, along with proteins, into the NCBI system. Once in the system nucleotides and proteins are both available for public use in at least three ways:
- 1.
The Entrez system (Chapter 15) retrieves nucleotide and protein sequences according to text queries that are entered into the search box. Text queries can be followed by search fields, such as author, definition line, and organism (for example, "homo sapiens"[orgn]), and are used to further define raw sequence data being used for retrieval.
- 2.
The sequences themselves can be searched directly by using BLAST (Chapter 16), which uses a sequence as a query to find similar sequences.
- 3.
Large subsets of sequences can be downloaded by FTP.
There are many sources for both nucleotide and protein sequences. Sequences submitted directly to GenBank (Chapter 1) or replicated from one of our two collaborating databases, the European Molecular Biology Laboratory (EMBL) Data Library and the DNA Data Bank of Japan (DDBJ), are the major sources. The Reference Sequence collection (Chapter 18) and the UniProt database, which incorporates data from SWISS-PROT, are yet additional sources.
An information management system that consists of two major components, the ID database and the IQ database, underlies the submission, storage, and access of GenBank, BLAST, and other curated data resources (such as the Reference Sequences (Chapter 18), the Map Viewer (Chapter 20), or Entrez Gene (Chapter 19)). Whereas ID handles incoming sequences and feeds other databases with subsets to suit different needs, IQ holds links between sequences stored in ID and between these sequences and other resources.
Abstract Syntax Notation 1 (ASN.1) Is the Data Format Used by the ID System
ASN.1 is the data description language in which all sequence data at NCBI are structured. ASN.1 allows a detailed description of both the sequences and the information associated with them, such as author names, source organism, and biological features (known as “features”). The image below shows FEATURES as displayed in GenBank format.

In the ASN.1 format, the organism information is presented as shown below. You can also see a complete ASN.1 record.
orgname { name binomial { genus "Macaca" , species "mulatta" } , Maintaining all data in the same structured format simplifies data parsing, manipulation, and quality assurance, and eases the task of data integration and software development for sequence analysis. All of the various divisions of GenBank can be downloaded in ASN.1 from the NCBI FTP site. In the ID data management system, data are stored as ASN.1 blobs, minimizing the amount of biological information that is captured and updated in the relational database schema.
Similar to an XML DTD, ASN.1 has an associated file that contains the description of the legal data structure. This file is called asn.all and is available as part of the “C” toolkit in an archive named “ncbi.tar.gz” located in the FTP directory. When unpacked, the directory “/demo”, found in the “ncbi.tar.gz” archive, contains the asn.all file. In the same “/demo” directory is testval.c, a tool that validates the data against asn.all. Additionally, a set of utilities for producing ASN.1 while programming in “C” is found in the subutil.c file of the “/api” directory, which is unpacked from the same “ncbi.tar.gz” archive.
Sources of Sequence Data
The sequence data available at NCBI comes from many different sources (Figure 1). In summary, the data consist of:

Figure
Figure 1. Sources of sequence data available at NCBI.
- Reference sequences (Chapter 18)
- sequences from the United States patents
The submission pathway depends on the data source (see Figure 1) and volume. HTGS and other large-volume submitters use FTP, usually after converting their data to ASN.1 with tools such as tabl2asn. Small-volume submitters typically use either BankIt (Chapter 1) or Sequin (Chapter 12) to prepare the ASN.1 for submission.
The data received are then subjected to some quality control by the submission tools BankIt, Sequin, and fa2htgs. These tools have built-in validation mechanisms to check if the data submitted have the correct structure and contain the essential information. The work of the GenBank indexing staff, who uses Sequin, adds one more layer of quality control and provides assistance to submitters. The staff also helps with the use of Sequin for complex submissions
Data Flow Components
The ID Database
The ID database is a group of standard relational databases that holds both ASN.1 objects and sequence identifier-related information. ASN.1 objects follow the specifications in the asn.all file for NCBI sequence data objects. ID holds data for GenBank and the many databases in the Entrez system. Details of the architecture of relational ID databases and the software associated with them are described later in this chapter. All of the sequences from the International Nucleotide Sequence Database Collaboration (INSDC)are in GenBank, and they all have Accession numbers assigned to them. Accession numbers point to sequences and their associated biological information and annotation.
In the ID database, blobs are added into a single column of a relational database. Although the columns behave as in a relational database, the information that makes each blob, such as biological features, raw sequence data, and author information, are neither parsed nor split out. In this sense, the ID database can be considered as a hybrid database that stores complex objects.
Note: Blob stands for Binary Large Object (or binary data object) and refers to a large piece of data, a large structured data object that can be stored as a unit and processed by software that knows the structure. For more information, check the Glossary.
Versions, GIs, Annotation Changes, and Takeovers
Every time a change is made to a sequence, a new version of the sequence is produced. This new version has a new GI number (GI or GenInfo Identifier is a sequence identification number for a nucleotide sequence) assigned to it (A and B in the image below). When a change is made to the annotation associated with a sequence, a new blob is produced, but no new version or GI is assigned. This series of events marks the history of the sequence since its first days in GenBank.
You can track annotation and sequence changes, as well as the “takeover” of one record by another by using the Sequence Revision History tool. The tool can be accessed from the side blue bar in Entrez Nucleotide and Entrez Protein and is used to highlight differences in sequence versions and annotations. To understand how the History tool works, let’s examine the history of the Gallus gallus doublesex and mab-3 related transcription factor 1 mRNA (Accession AF123456), which was first added to GenBank March 20, 1999.
Click on Check sequence revision history in the blue side bar of Entrez Nucleotide or Entrez Protein to be directed to the Sequence Revision History page. Enter the Accession or GI numbers or the FASTA-style Sequence IDs (SeqIds) into the Find box. The Revision history for AF123456 is displayed.
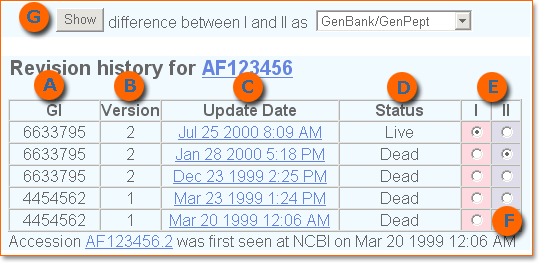
The Update Date column (C in the image above) contains the date of every update to AF123456. Some involve sequence changes, others involve only annotation changes. Click on a date in the column to retrieve AF123456 as it existed at that point in time. The status column (D) reports which version is live and which ones are dead. Columns I and II (E) are used to compare two different sequences.
Notice that on Mar 23 1999, at 1:24 PM, a new ASN.1 blob was produced for Accession AF12345. However, no new GI number (A) or version (B) was assigned because the changes were limited to the annotation and biological features of the sequence, with no changes made to the sequence data. On December 23, 1999, Accession AF123456 gained a new GI (6633795) and version (Version 2) because in this case a change was made to the sequence data.
Compare the two blobs produced on March 23, 1999 and December 23, 1999 to see the difference between them.
- Select one sequence in each column (I or II) as shown in the image above (E).
- Push the Show button at the upper left of the page to display the two blobs (G).
The differences between blobs are highlighted, with each blob displaying a different color. Compare ASN.1 blobs produced on March 20, 1999 and March 23, 1999 and you will see that the differences between the two are limited to the annotation and biological features described in the blobs, whereas the sequence data remain the same.
The understanding of the biological features related to a sequence can change with or without a change in the underlying genetic sequence. For example, the sequence revision history of J00179 reveals that although the annotation changed four times, there has been only one sequence version (J00179) with one GI (183807). J00179 can still be retrieved in Entrez by searching its Accession or GI number, but this record has been replaced by Accession U01317 and therefore is no longer indexed. The version number assigned to the “take over” record U01317 is 1, whereas the replaced version of this record (J00179) remains as Version 0. All sequences deposited before February 1999 received no sequence version, that’s why J00179 is version zero. In February 1999, the use of a sequence version was implemented, and all sequences deposited in GenBank at that time received a version number 1. Since then, ordinals assigned to sequence versions have increased every time a change is made to the sequence data.
The use of both systems, Version and GI, leads to two parallel ways of tracking sequence versions for an object. In the GenBank flatfile, the Accession Version provides the ordinal instance (version) of the sequence. Within ID, each unique sequence is assigned a GI number; and therefore the instances of an Accession can be tracked by checking its chain of GI numbers. Note that Accession and Accession Version are different things, with the former been used to designate a DNA sequence of some molecule or piece of some molecule deposited in GenBank and the latter to indicate the version of that sequence. A single Accession can have many GIs that are assigned every time the sequence changes, whereas an Accession Version has only one GI.
Within the ID relational databases, there is a chain identifier that can be used to link these GI numbers. Not all sequences within ID are in GenBank and not all have sequence versions, but all sequences have a chain of GI numbers. For this reason, internally, the GI number is the universal pointer to a particular sequence, as opposed to the Accession Version, which would work only for versioned sequences. The ID database is also the controller for allowed “takeovers” of one Accession by another. In the example above, GI 4454562 is taken over by GI 6633795. A takeover can also occur when the sequences of two clones are merged into a single clone. One or several of the Accessions of older clones can be taken over by a new Accession.
Output of Data from the ID System
Once all incoming data have been converted to ASN.1 format and entered into ID, the data are then replicated into several different servers and transformed into several different formats (Figure 2). The replication is necessary for a number of reasons: (i) it separates the “incoming” data system (ID) from the “outgoing” data which is the data used in response to scientific queries by users; (ii) it helps balance the load of queries, thus providing quicker response times and allowing different servers to specialize in different functions; and (iii) it protects against data loss should one server fail. The details of the internal structure of the ID system and how the structure is replicated are discussed in the Data Flow Architecture section.
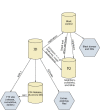
Figure
Figure 2. Products of the ID system.
The IQ Database
The IQ database is a Sybase data-warehousing product that preserves its SQL language interface but which inverts its data by storing it by column, not by row. Its strength is in its ability to speed up results from queries based on the anticipated indexing. This non-relational database holds links between many different objects.
For example, as part of the processing of incoming sequences, each protein and nucleotide sequence is searched for similar sequences (Chapter 16) against the rest of the database. Users can then select the Related Sequences link that is displayed next to each record in Entrez Nucleotide and Entrez Protein (Chapter 15) to see a set of similar sequences, sometimes known as “neighbors”. The IQ database keeps track of the neighbors for any given sequence. These relationships are all pre-computed to save users’ time.
IQ stores the relationships between similar nucleotide sequences and between similar protein sequences and which proteins are coded for by which nucleotides and also holds information on the links between entries in different Entrez databases. This might include, for example, information on the publications cited within sequence records, which links to PubMed or to an organism in the Taxonomy database. Some of this information comes from the analysis of the ASN.1 in ID by e2index, a tool that extracts terms from NCBI sequence ASN.1 during “indexing” for Entrez.
The BLAST Control Database
The BLAST Control database receives information from ID that is used to generate BLAST databases (Chapter 16) for the BLAST query service and for stand-alone BLAST users. The information is used internally to generate the sequence neighbors stored in IQ.
The GenBank Flatfile and Error Capture Databases
Many NCBI users think of the GenBank flatfile as the archetypal sequence data format (see an example of a GenBank flatfile). However, within NCBI and especially within the ID internal data flow system, ASN.1 is considered the original format from which reports such as the GenBank flatfile can be generated (see an example of an ASN.1 file).
Although the GenBank flatfile is usually generated on demand from the ASN.1, for certain products such as complete GenBank releases, a GenBank flatfile image is made for each active sequence. This flatfile is stored in a database called FF4Release, which consists of the latest transformation of ASN.1 to the GenBank flatfile format.
The FF4Release database is also a place where internal error reports are captured. The reports can be analyzed and displayed for different time points in the data processing pathway:
Entrez Postings Files
When sequences are submitted to GenBank or one of our collaborating databases, additional information about the sequence is often included. This might be a brief description of a gene in the definition line, along with annotated sequence features such as the source organism name. To make this information searchable via Entrez, these words have to be indexed. They are extracted from the ASN.1 using e2index and then stored in the Entrez posting files, which are optimized for Boolean queries by the Entrez system (see Chapter 15).
All of these products from the ID system are listed in Table 1. NCBI also generates weekly “LiveLists” for public, collaborator, and in-house use. LiveLists show all Accession numbers currently in use. Accession numbers that have been replaced or otherwise removed from circulation because of error or submitter request are not in the LiveList.
Table
Table 1. Products of the ID system.
Data Flow Architecture
Sequences enter ID when a client (internal to NCBI) loads data into the system. The ASN.1 data can be loaded either through a stand-alone program or a client API. In both cases, the data are submitted to ID through IDProdOS, an open server (commonly called “middleware”) that sits between the clients and the database system. An overview of the flow of sequence data through the ID architecture with its multiple components is shown in Figure 3 and discussed below.
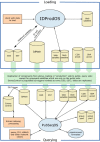
Figure
Figure 3. The ID system architecture.
IDProdOS hides details of the underlying complexity from the client API, which was shown to be useful when the previous version of the ID system (a single database and an open server) was converted to the current system without requiring any changes to the clients.
IDProdOS does an initial check of the actions required by the load. For example, in a record that has DNA and protein sequences, including annotation and sequence identifiers, the identifier on the protein has to be unique. The same identifier should not be given to an outdated DNA sequence and a current sequence, unless the current sequence has replaced the old one. That’s because proteins, generally, are not allowed to move between GenBank records, although proteins moving between segments of a complete genome submission are sometimes allowed.
Additional checking is performed by stored procedures in the IdMain database. The details of what is allowed vary according to the source of the ASN.1, which includes direct submissions from collaborators and the NCBI RefSeq project. These procedures check (i) which sequence identifiers may be used, (ii) which sequences may be replaced by which other sequences, and (iii) which sequence version may be used in a record.
If the sequences pass all these checks, three things happen: (i) IDProdOS changes the SeqId pointers in the blob to GI numbers, which are now used as sequence-specific pointers, (ii) IdMain retains the sequence identifier information that was also used for the checking, and (iii) IDProdOS loads the ASN.1 blobs to the blob satellites.
The IdMain database contains the sequence identifiers for each of the sequence records, including all those for ASN.1 blobs that contain multiple sequences. It enforces sequence version rules, among other rules.
Relational satellite databases are fully normalized databases that hold records for which there is only one sequence per intended ASN.1 blob. Few, if any, features are allowed on records intended for relational satellite databases (the PubSeqOS produces the ASN.1 by converting the data extracted from relational tables). This contrasts with the Blob satellite databases, from which ASN.1 is retrieved as-is. Blob satellite databases, different from relational databases, contain ASN.1 objects as unnormalized data objects.
Recently, annotation-only satellite databases have been added to the ID system. These satellites contain annotation to be added to Bioseqs, linked by GI number. Because there are multiple such annotation satellite databases, more than one set of additional annotation may be added to a Bioseq.
The SnpAnnot database contains feature information that is limited to simple mutation information from dbSNP (Chapter 5). The CDD Annotation database contains feature information that is limited to protein domains for the protein sequences known to ID. In both cases, these features might be added to NCBI-curated records by the PubSeqOS when the records are requested.
To visualize the role of replication, the rectangle in the middle of Figure 3 represents the use of the Sybase Replication Server to copy information from the loading side of the system to the query side.
Similar to IDProdOS, PubSeqOS is a open server (also called “middleware”) that sits between the clients and the database system. It hides details of the underlying complexity from the client API. It actually has an almost identical code base as IDProdOS because they both serve similar functions. When a record is requested in a format other than ASN.1, psansconvert is called to do the conversion. This distinct child process allows both insulation from any possible instability and allows for use of multiple central processing units (CPUs) in a natural way.
Note: The child process is a technical term used to describe a process that is owned by and completely dependent on a parent process that initiated it.
At the query side are all records in Entrez, plus graveyards and EntrezControl, a special database that is not queried by the public. EntrezControl is used to control the indexing of blobs for Entrez. Its rows are initiated by a trigger that fires when rows are added by replication to the IdMan database. A trigger is a special, database-stored procedure that responds to changes in a database table.
The graveyards are databases that contain blobs that were replaced or taken over and therefore no longer indexed in Entrez. Once replaced or taken over, blobs do not change—which is the reason why they are limited to the query side—but they are still retrievable by GI or other sequence identifier.
- The Processing of Biological Sequence Data at NCBI - The NCBI HandbookThe Processing of Biological Sequence Data at NCBI - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...

