| NCBI National Center for Biotechnology Information |  |
2X80:
P450 BM3 F87A in complex with DMSO
| Biological unit 1: | monomeric |
| Source organism: | Priestia megaterium |
| Number of proteins: | 1 (BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE) |
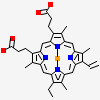
| Number of chemicals: | 3 (DIMETHYL SULFOXIDE,PROTOPORPHYRIN IX CONTAINING... ▼) Chemical
close |
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Full |
4HGI | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) |
Priestia megaterium |
1 | 0.56Å | 455 | 99% | |
| 2 | Full |
4HGH | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) |
Priestia megaterium |
1 | 0.57Å | 455 | 99% | |
| 3 | Full |
6JLV | Near-Atomic Resolution Structure of the CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan |
Priestia megaterium |
1 | 0.60Å | 455 | 99% | |
| 4 | Full |
1ZO4 | Crystal Structure Of A328S Mutant Of The Heme Domain Of P450BM-3 |
Priestia megaterium |
1 | 0.61Å | 455 | 99% | |
| 5 | Full |
4DUA | cytochrome P450 BM3h-9D7 MRI sensor, no ligand |
Priestia megaterium |
1 | 0.66Å | 455 | 98% | |
| 6 | Full |
2HPD | CRYSTAL STRUCTURE OF HEMOPROTEIN DOMAIN OF P450BM-3, A PROTOTYPE FOR MICROSOMAL P450'S |
Priestia megaterium |
1 | 0.69Å | 455 | 99% | |
| 7 | Full |
6JMW | Structure of the Chromium Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan |
Priestia megaterium |
1 | 0.72Å | 455 | 99% | |
| 8 | Full |
6JS8 | Structure of the CYP102A1 Haem Domain with N-Dehydroabietoyl-L-Tryptophan |
Priestia megaterium |
1 | 0.74Å | 455 | 99% | |
| 9 | Full |
6JMH | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan |
Priestia megaterium |
1 | 0.74Å | 455 | 99% | |
| 10 | Full |
6K58 | Structure of the CYP102A1 Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine |
Priestia megaterium |
1 | 0.76Å | 455 | 99% |