| NCBI National Center for Biotechnology Information |  |
3DVA:
Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex
| Biological unit 1: | pentameric | ||||||
| Source organism: | Geobacillus stearothermophilus | ||||||
| Number of proteins: | 5 (Dihydrolipoyllysine-residue acetyltransferase c... ▼) Protein molecule
close
|
||||||
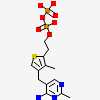
| Number of chemicals: | 7 (POTASSIUM ION (2),MAGNESIUM ION (3),2-{4-[(4-AM... ▼)
|
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Partial |
3DUF | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex |
Others |
4 | 0.46Å | 1378 | 99% | |
| 2 | Partial |
1W85 | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 |
Geobacillus stearothermophilus |
4 | 0.31Å | 1371 | 99% | |
| 3 | Partial |
3DV0 | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex |
Others |
4 | 0.27Å | 1347 | 99% | |
| 4 | Partial |
1UMD | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate |
Thermus thermophilus |
4 | 1.57Å | 1337 | 45% | |
| 5 | Partial |
1UMB | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form |
Thermus thermophilus |
4 | 1.57Å | 1337 | 45% | |
| 6 | Partial |
1UMC | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methylpentanoate |
Thermus thermophilus |
4 | 1.62Å | 1337 | 45% | |
| 7 | Partial |
1DTW | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE |
Homo sapiens |
4 | 1.92Å | 1336 | 35% | |
| 8 | Partial |
2BFF | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch |
Homo sapiens |
4 | 1.82Å | 1333 | 36% | |
| 9 | Partial |
1W88 | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 |
Geobacillus stearothermophilus |
4 | 0.37Å | 1331 | 99% | |
| 10 | Partial |
1U5B | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase |
Homo sapiens |
4 | 1.79Å | 1330 | 36% |