| NCBI National Center for Biotechnology Information |  |
2BR5:
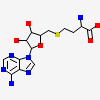
cmcI-N160 SAH
| Biological unit 1: | hexameric | ||
| Source organism: | Streptomyces clavuligerus | ||
| Number of proteins: | 6 (CEPHALOSPORIN HYDROXYLASE CMCI ▼) Protein molecule
close
|
||
| Number of chemicals: | 5 (S-ADENOSYL-L-HOMOCYSTEINE (5) ▼) Chemical
close |
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Full |
2BM9 | cmcI-N160 in complex with SAM |
Streptomyces clavuligerus |
6 | 0.81Å | 1256 | 100% | |
| 2 | Full |
2BR4 | cmcI-D160 Mg-SAM |
Streptomyces clavuligerus |
6 | 0.63Å | 1255 | 99% | |
| 3 | Full |
2BR3 | cmcI-D160 Mg |
Streptomyces clavuligerus |
6 | 0.79Å | 1253 | 99% | |
| 4 | Full |
2BM8 | Cmci-N160 Apo-Structure |
Streptomyces clavuligerus |
6 | 0.79Å | 1250 | 100% | |
| 5 | Full |
7ULH | Crystal Structure of a Short chain dehydrogenase from Mycobacterium avium 104 |
Mycobacterium avium 104 |
6 | 18.95Å | 595 | 12% | |
| 6 | Partial |
3PXX | Crystal structure of carveol dehydrogenase from Mycobacterium avium bound to nicotinamide adenine dinucleotide |
Mycobacterium avium 104 |
3 | 5.81Å | 297 | 13% | |
| 7 | Partial |
5CCX | Structure of the product complex of tRNA m1A58 methyltransferase with tRNA3Lys as substrate |
Homo sapiens |
2 | 5.96Å | 272 | 11% | |
| 8 | Partial |
2YVL | Crystal structure of tRNA (m1A58) methyltransferase TrmI from Aquifex aeolicus |
Aquifex aeolicus VF5 |
2 | 4.88Å | 263 | 11% | |
| 9 | Partial |
3NJR | Crystal structure of C-terminal domain of precorrin-6Y C5,15-methyltransferase from Rhodobacter capsulatus |
Rhodobacter capsulatus SB 1003 |
2 | 5.41Å | 261 | 11% | |
| 10 | Partial |
3NJR | Crystal structure of C-terminal domain of precorrin-6Y C5,15-methyltransferase from Rhodobacter capsulatus |
Rhodobacter capsulatus SB 1003 |
2 | 5.41Å | 261 | 11% |