| NCBI National Center for Biotechnology Information |  |
4M11:
Crystal Structure of Murine Cyclooxygenase-2 Complex with Meloxicam
| Biological unit 1: | dimeric | ||
| Source organism: | Mus musculus | ||
| Number of proteins: | 2 (Prostaglandin G/H synthase 2 ▼) Protein molecule
close
|
||
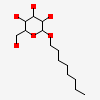
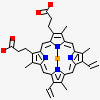
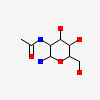
| Number of chemicals: | 17 (octyl beta-D-glucopyranoside,PROTOPORPHYRIN IX ... ▼)
|
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Full |
4M10 | Crystal Structure of Murine Cyclooxygenase-2 Complex with Isoxicam |
Mus musculus |
2 | 0.19Å | 1104 | 100% | |
| 2 | Full |
4RUT | crystal structure of murine cyclooxygenase-2 with 13-methyl-arachidonic Acid |
Mus musculus |
2 | 0.24Å | 1104 | 100% | |
| 3 | Full |
3LN1 | Structure Of Celecoxib Bound At The Cox-2 Active Site |
Mus musculus |
2 | 0.29Å | 1104 | 100% | |
| 4 | Full |
3LN0 | Structure Of Compound 5c-S Bound At The Active Site Of Cox-2 |
Mus musculus |
2 | 0.32Å | 1104 | 100% | |
| 5 | Full |
6BL4 | Crystal Complex of Cyclooxygenase-2 with indomethacin-ethylenediamine-dansyl conjugate |
Mus musculus |
2 | 0.34Å | 1104 | 100% | |
| 6 | Full |
6BL3 | Crystal Complex of Cyclooxygenase-2 with indomethacin-butyldiamine-dansyl conjugate |
Mus musculus |
2 | 0.35Å | 1104 | 100% | |
| 7 | Full |
4RRY | Crystal Structure of Apo Murine H90W Cyclooxygenase-2 |
Mus musculus |
2 | 0.35Å | 1104 | 99% | |
| 8 | Full |
4RRW | Crystal Structure of Apo Murine Cyclooxygenase-2 |
Mus musculus |
2 | 0.35Å | 1104 | 99% | |
| 9 | Full |
4RRZ | Crystal Structure of Apo Murine H90W Cyclooxygenase-2 Complexed with Lumiracoxib |
Mus musculus |
2 | 0.35Å | 1104 | 99% | |
| 10 | Full |
3RR3 | Structure Of (R)-Flurbiprofen Bound To Mcox-2 |
Mus musculus |
2 | 0.37Å | 1104 | 100% |